2E6X
 
 | | X-ray structure of TT1592 from Thermus thermophilus HB8 | | Descriptor: | Hypothetical protein TTHA1281 | | Authors: | Yamada, M, Nakagawa, N, Kanagawa, M, Kuramitsu, S, Kamitori, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of TT1592 from Thermus thermophilus HB8
To be Published
|
|
1WZK
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt D465N | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZL
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZM
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469K | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1UG9
 
 | | Crystal Structure of Glucodextranase from Arthrobacter globiformis I42 | | Descriptor: | CALCIUM ION, GLYCEROL, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Ohtaki, A, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1VFO
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/beta-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
2ZYO
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with maltotetraose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
2ZYM
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
1VFM
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/alpha-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1VFU
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 amylase 2/gamma-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
2ZYN
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with beta-cyclodextrin | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
3A9T
 
 | | X-ray Structure of Bacillus pallidus D-Arabinose Isomerase Complex with L-Fucitol | | Descriptor: | D-arabinose isomerase, FUCITOL, MANGANESE (II) ION | | Authors: | Takeda, K, Yoshida, H, Izumori, K, Kamitori, S. | | Deposit date: | 2009-11-05 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | X-ray structures of Bacillus pallidusd-arabinose isomerase and its complex with l-fucitol.
Biochim.Biophys.Acta, 1804, 2010
|
|
3ALM
 
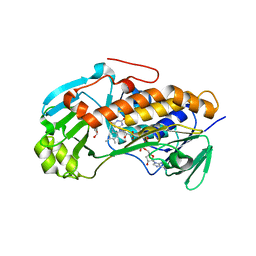 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant C294A | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
3A9S
 
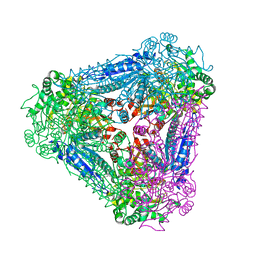 | | X-ray Structure of Bacillus pallidus D-Arabinose Isomerase Complex with Glycerol | | Descriptor: | D-arabinose isomerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Takeda, K, Yoshida, H, Izumori, K, Kamitori, S. | | Deposit date: | 2009-11-05 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structures of Bacillus pallidusd-arabinose isomerase and its complex with l-fucitol.
Biochim.Biophys.Acta, 1804, 2010
|
|
1ULV
 
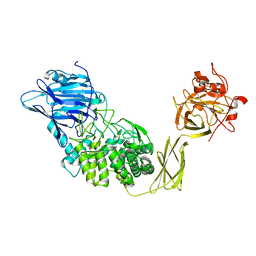 | | Crystal Structure of Glucodextranase Complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
3A6O
 
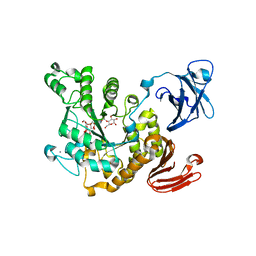 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/acarbose complex | | Descriptor: | ACARBOSE DERIVED PENTASACCHARIDE, CALCIUM ION, Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2009-09-07 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
3A9R
 
 | | X-ray Structures of Bacillus pallidus D-Arabinose IsomeraseComplex with (4R)-2-METHYLPENTANE-2,4-DIOL | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, D-arabinose isomerase, MANGANESE (II) ION | | Authors: | Takeda, K, Yoshida, H, Izumori, K, Kamitori, S. | | Deposit date: | 2009-11-05 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | X-ray structures of Bacillus pallidusd-arabinose isomerase and its complex with l-fucitol.
Biochim.Biophys.Acta, 1804, 2010
|
|
3W7T
 
 | | Escherichia coli K12 YgjK complexed with mannose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Uncharacterized protein YgjK, ... | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
3W7S
 
 | | Escherichia coli K12 YgjK complexed with glucose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-glucopyranose | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
4H2R
 
 | | Structure of MHPCO Y270F mutant, 5-hydroxynicotinic acid complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
2ECE
 
 | |
3WUC
 
 | | X-ray crystal structure of Xenopus laevis galectin-Va | | Descriptor: | Galectin, MALONIC ACID, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nonaka, Y, Yoshida, H, Kamitori, S, Nakamura, T. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Xenopus laevis skin proto-type galectin, close to but distinct from galectin-1.
Glycobiology, 25, 2015
|
|
3WW3
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with no ligand | | Descriptor: | L-ribose isomerase, MANGANESE (II) ION | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4GF7
 
 | | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Sawa, Y, Yagi, T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
3WW1
 
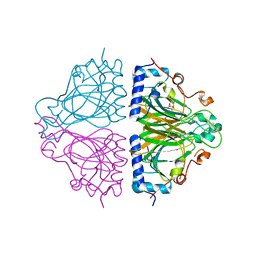 | | X-ray structure of Cellulomonas parahominis L-ribose isomerase with L-ribose | | Descriptor: | L-ribose, L-ribose isomerase, MANGANESE (II) ION, ... | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
