6MW8
 
 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Manganese | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
1BLL
 
 | |
1BPM
 
 | |
7X15
 
 | | Crystal structure of MIGA2 LD targeting domain | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, FORMIC ACID, Mitoguardin 2 | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
7X14
 
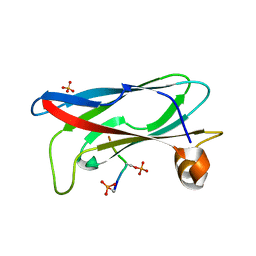 | | Crystal structure of phospho-FFAT motif of MIGA2 bound to VAPB | | Descriptor: | MIGA2 phospho FFAT motif, SULFATE ION, Vesicle-associated membrane protein-associated protein B | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
4DXO
 
 | |
4DXN
 
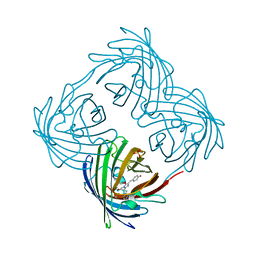 | |
4DXP
 
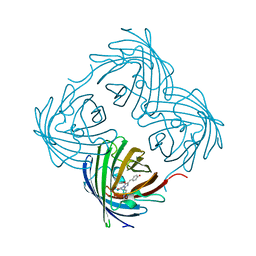 | |
4DXQ
 
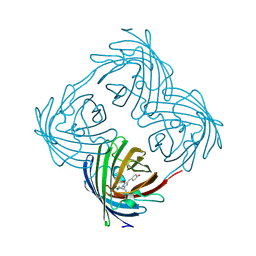 | |
3PG4
 
 | |
5YU2
 
 | | Structure of Ribonuclease YabJ | | Descriptor: | Translation initiation inhibitor homologue | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-11-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel chlorination-induced ribonuclease YabJ fromStaphylococcus aureus.
Biosci. Rep., 38, 2018
|
|
6MW5
 
 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Platinum | | Descriptor: | 1,2-ETHANEDIOL, PLATINUM (II) ION, UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
6KHM
 
 | | Lipase (Open form) | | Descriptor: | Hydrolase, alpha/beta domain protein, heptane-1,1-diol, ... | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Closed, blocked, and open states of lipase from type II Cutibacterium acnes
To Be Published
|
|
3W6M
 
 | |
3W6L
 
 | |
1J32
 
 | |
4Q2T
 
 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-arginine | | Descriptor: | ARGININE, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
1GH6
 
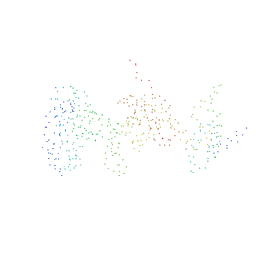 | | RETINOBLASTOMA POCKET COMPLEXED WITH SV40 LARGE T ANTIGEN | | Descriptor: | Large T antigen, Retinoblastoma-associated protein | | Authors: | Kim, H.Y, Cho, Y. | | Deposit date: | 2000-11-15 | | Release date: | 2001-11-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen.
EMBO J., 20, 2001
|
|
6KHK
 
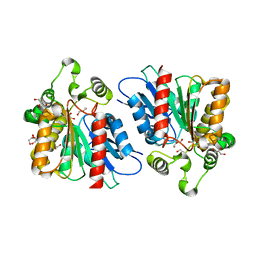 | | Lipase (Closed form) | | Descriptor: | GLYCEROL, Hydrolase, alpha/beta domain protein | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Closed, blocked, and open states of lysophospholipase from type II Cutibacterium acnes
To Be Published
|
|
6KHL
 
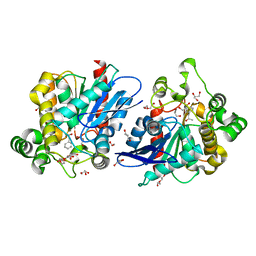 | | Lipase (Blocked form) | | Descriptor: | (2~{R})-1-phenylpropan-2-ol, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Closed, blocked, and open states of lipase from type II Cutibacterium acnes
To Be Published
|
|
4Q2Y
 
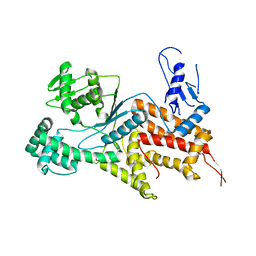 | | Crystal structure of Arginyl-tRNA synthetase | | Descriptor: | Arginine--tRNA ligase, cytoplasmic | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
4Q2X
 
 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-canavanine | | Descriptor: | Arginine--tRNA ligase, cytoplasmic, L-CANAVANINE | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
3OLM
 
 | | Structure and Function of a Ubiquitin Binding Site within the Catalytic Domain of a HECT Ubiquitin Ligase | | Descriptor: | E3 ubiquitin-protein ligase RSP5, Ubiquitin | | Authors: | Kim, H.C, Steffen, A, Chen, J, Huibregtse, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structure and function of a HECT domain ubiquitin-binding site.
Embo Rep., 12, 2011
|
|
7CTO
 
 | | Staphylococcus aureus MsrB | | Descriptor: | Peptide methionine sulfoxide reductase MsrB | | Authors: | Kim, H.J. | | Deposit date: | 2020-08-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Implication of Staphylococcus aureus MsrB dimerization upon oxidation.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
8HEJ
 
 | | Crystal structure of Transthyretin in complex with a covalent inhibitor trans-styrylpyrazole | | Descriptor: | 2,4,6-trifluorobenzaldehyde, 2,6-dibromo-4-[(E)-2-(3,5-dimethyl-1H-pyrazol-4-yl)ethenyl]phenol, Transthyretin | | Authors: | Kim, H, Choi, S, Lee, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of Transthyretin in complex with a covalent inhibitor trans-styrylpyrazole
To Be Published
|
|
