2FZT
 
 | |
2ITB
 
 | |
2FTR
 
 | |
2FMU
 
 | |
2II1
 
 | |
4Q5K
 
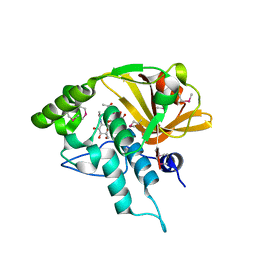 | | Crystal structure of a N-acetylmuramoyl-L-alanine amidase (BACUNI_02947) from Bacteroides uniformis ATCC 8492 at 1.30 A resolution | | Descriptor: | (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid, SODIUM ION, Uncharacterized protein | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-guided functional characterization of DUF1460 reveals a highly specific NlpC/P60 amidase family.
Structure, 22, 2014
|
|
2GC9
 
 | |
2GA1
 
 | |
2GF6
 
 | |
2GI3
 
 | |
2G8L
 
 | |
2G80
 
 | |
2FVG
 
 | |
2FYX
 
 | |
2G0T
 
 | |
2G0W
 
 | |
2FTZ
 
 | |
4Q68
 
 | |
2GJG
 
 | |
2H0V
 
 | |
2HI0
 
 | |
2B8N
 
 | |
2HSB
 
 | |
2HUH
 
 | |
2HR2
 
 | |
