1J5N
 
 | | Solution Structure of the Non-Sequence-Specific HMGB protein NHP6A in complex with SRY DNA | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*CP*AP*AP*TP*CP*AP*CP*CP*CP*C)-3', 5'-D(*GP*GP*GP*GP*TP*GP*AP*TP*TP*GP*TP*TP*CP*AP*G)-3', Nonhistone chromosomal protein 6A | | Authors: | Masse, J.E, Wong, B, Yen, Y.-M, Allain, F.H.-T, Johnson, R.C, Feigon, J. | | Deposit date: | 2002-05-15 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding
J.Mol.Biol., 323, 2002
|
|
1LWM
 
 | | Solution Structure of the Sequence-Non-Specific HMGB protein NHP6A | | Descriptor: | NONHISTONE CHROMOSOMAL PROTEIN 6A | | Authors: | Masse, J.E, Wong, B, Yen, Y.-M, Allain, F.H.-T, Johnson, R.C, Feigon, J. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding
J.Mol.Biol., 323, 2002
|
|
1LWA
 
 | | Solution Structure of SRY_DNA | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*CP*AP*AP*TP*CP*AP*CP*CP*CP*C)-3', 5'-D(*GP*GP*GP*GP*TP*GP*AP*TP*TP*GP*TP*TP*CP*AP*G)-3' | | Authors: | Masse, J.E, Wong, B, Yen, Y.-M, Allain, F.H.-T, Johnson, R.C, Feigon, J. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding
J.Mol.Biol., 323, 2002
|
|
1ETO
 
 | |
1ETQ
 
 | |
1ETV
 
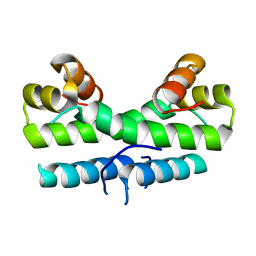 | |
1FIP
 
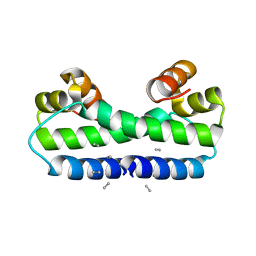 | | THE STRUCTURE OF FIS MUTANT PRO61ALA ILLUSTRATES THAT THE KINK WITHIN THE LONG ALPHA-HELIX IS NOT DUE TO THE PRESENCE OF THE PROLINE RESIDUE | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS), UNKNOWN PEPTIDE, POSSIBLY PART OF THE UNOBSERVED RESIDUES IN ENTITY 1 | | Authors: | Yuan, H.S, Wang, S.S, Yang, W.-Z, Finkel, S.E, Johnson, R.C. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of Fis mutant Pro61Ala illustrates that the kink within the long alpha-helix is not due to the presence of the proline residue.
J.Biol.Chem., 269, 1994
|
|
1CG7
 
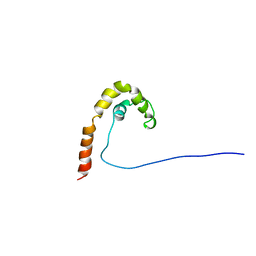 | | HMG PROTEIN NHP6A FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PROTEIN (NON HISTONE PROTEIN 6 A) | | Authors: | Allain, F.H.T, Yen, Y.M, Masse, J.E, Schultze, P, Dieckmann, T, Johnson, R.C, Feigon, J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding.
EMBO J., 18, 1999
|
|
1ETK
 
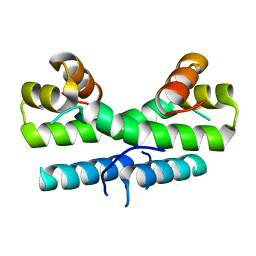 | |
1ETY
 
 | |
1ETX
 
 | |
1ETW
 
 | |
3FIS
 
 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1RH6
 
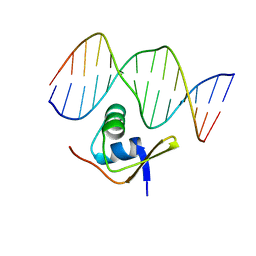 | | Bacteriophage Lambda Excisionase (Xis)-DNA Complex | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*TP*AP*GP*TP*CP*TP*GP*TP*TP*G)-3', 5'-D(P*CP*AP*AP*CP*AP*GP*AP*CP*TP*AP*CP*AP*TP*AP*G)-3', Excisionase | | Authors: | Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the excisionase-DNA complex from bacteriophage lambda.
J.Mol.Biol., 338, 2004
|
|
2IEF
 
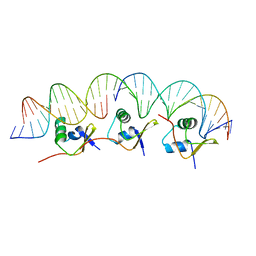 | | Structure of the cooperative Excisionase (Xis)-DNA complex reveals a micronucleoprotein filament | | Descriptor: | 15-mer DNA, 19-mer DNA, 34-mer DNA, ... | | Authors: | Abbani, M.A, Papagiannis, C.V, Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2006-09-18 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3IV5
 
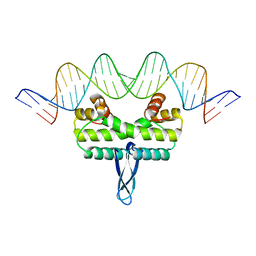 | |
2OG0
 
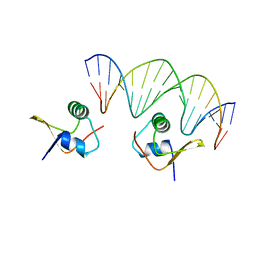 | | Crystal Structure of the Lambda Xis-DNA complex | | Descriptor: | 5'-D(*AP*AP*AP*CP*AP*GP*AP*CP*TP*AP*CP*AP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*TP*AP*TP*TP*AP*TP*GP*TP*AP*GP*TP*CP*TP*GP*TP*TP*T)-3', Excisionase | | Authors: | Papagiannis, C.V, Sam, M.D, Abbani, M.A, Cascio, D, Yoo, D, Clubb, R.T, Johnson, R.C. | | Deposit date: | 2007-01-04 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fis targets assembly of the xis nucleoprotein filament to promote excisive recombination by phage lambda.
J.Mol.Biol., 367, 2007
|
|
3JR9
 
 | |
3JRF
 
 | |
3JRD
 
 | |
3JRB
 
 | |
3JRH
 
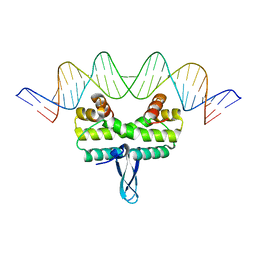 | |
3JRA
 
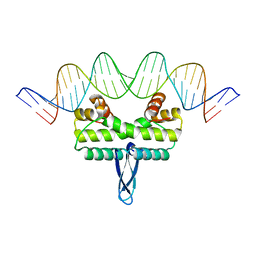 | |
3JRC
 
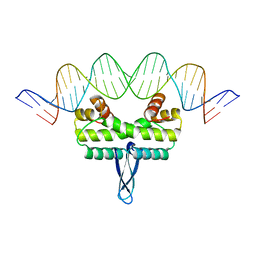 | |
3JRI
 
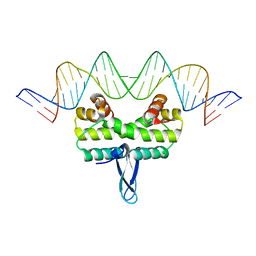 | |
