2W04
 
 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with citrate in ATP binding site from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, CITRATE ANION | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2W8J
 
 | | SPT with PLP-ser | | Descriptor: | SERINE PALMITOYLTRANSFERASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Carter, L.G, Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-16 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
2W8T
 
 | | SPT with PLP, N100C | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-19 | | Release date: | 2009-01-27 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
1VJI
 
 | | Gene Product of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase (OPR1), FLAVIN MONONUCLEOTIDE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1.
Proteins, 61, 2005
|
|
1VK5
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, SULFATE ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structure at 1.6 A resolution of the protein from gene locus At3g22680 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VYF
 
 | | schistosoma mansoni fatty acid binding protein in complex with oleic acid | | Descriptor: | 14 KDA FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Angelucci, F, Johnson, K.A, Baiocco, P, Miele, A.E, Bellelli, A. | | Deposit date: | 2004-04-29 | | Release date: | 2004-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Schistosoma Mansoni Fatty Acid Binding Protein: Specificity and Functional Control as Revealed by Crystallographic Structure
Biochemistry, 43, 2004
|
|
1VYG
 
 | | schistosoma mansoni fatty acid binding protein in complex with arachidonic acid | | Descriptor: | ARACHIDONIC ACID, FATTY ACID BINDING PROTEIN | | Authors: | Angelucci, F, Johnson, K.A, Baiocco, P, Miele, A.E, Bellelli, A. | | Deposit date: | 2004-04-29 | | Release date: | 2004-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Schistosoma Mansoni Fatty Acid Binding Protein: Specificity and Functional Control as Revealed by Crystallographic Structure
Biochemistry, 43, 2004
|
|
2IX2
 
 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2JGR
 
 | | Crystal structure of YegS in complex with ADP | | Descriptor: | PYROPHOSPHATE 2-, YEGS | | Authors: | Bakali, H.M, Herman, M.D, Johnson, K.A, Kelly, A.A, Wieslander, A, Hallberg, B.M, Nordlund, P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Yegs, a Homologue to the Mammalian Diacylglycerol Kinases, Reveals a Novel Regulatory Metal Binding Site.
J.Biol.Chem., 282, 2007
|
|
2JG2
 
 | | HIGH RESOLUTION STRUCTURE OF SPT WITH PLP INTERNAL ALDIMINE | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-05-01 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
2JG5
 
 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG6
 
 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JGT
 
 | | Low resolution structure of SPT | | Descriptor: | SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-14 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
2FGE
 
 | |
2BON
 
 | | Structure of an Escherichia coli lipid kinase (YegS) | | Descriptor: | LIPID KINASE, MAGNESIUM ION | | Authors: | Bakali, H.M, Johnson, K.A, Hallberg, B.M, Herman, M.D, Nordlund, P. | | Deposit date: | 2005-04-12 | | Release date: | 2006-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Yegs, a Homologue to the Mammalian Diacylglycerol Kinases, Reveals a Novel Regulatory Metal Binding Site.
J.Biol.Chem., 282, 2007
|
|
2FRX
 
 | | Crystal structure of YebU, a m5C RNA methyltransferase from E.coli | | Descriptor: | Hypothetical protein yebU | | Authors: | Erlandsen, H, Nordlund, P, Hallberg, B.M, Johnson, K.A, Ericsson, U.B. | | Deposit date: | 2006-01-20 | | Release date: | 2006-08-29 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain
J.Mol.Biol., 360, 2006
|
|
6ENK
 
 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
1ZWJ
 
 | | X-ray structure of galt-like protein from arabidopsis thaliana AT5G18200 | | Descriptor: | ZINC ION, putative galactose-1-phosphate uridyl transferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, McCoy, J.G, Johnson, K.A, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of an ADP-Glucose Phosphorylase from
Arabidopsis thaliana
Biochemistry, 45, 2006
|
|
1VK0
 
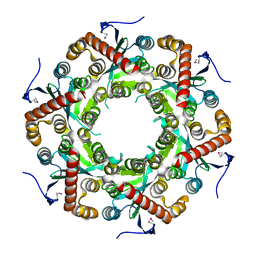 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450 | | Descriptor: | hypothetical protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450
To be published
|
|
2WR8
 
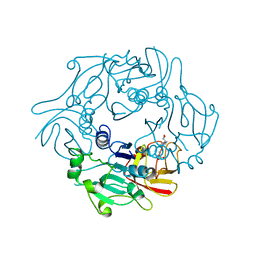 | | Structure of Pyrococcus horikoshii SAM hydroxide adenosyltransferase in complex with SAH | | Descriptor: | PUTATIVE UNCHARACTERIZED PROTEIN PH0463, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMahon, S.A, Deng, H, O'Hagan, D, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Mechanistic Insights Into Water Activation in Sam Hydroxide Adenosyltransferase (Duf-62).
Chembiochem, 10, 2009
|
|
2VV5
 
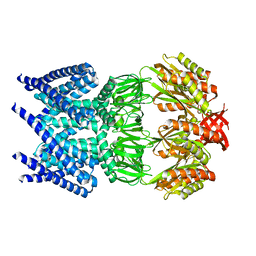 | | The open structure of MscS | | Descriptor: | SMALL-CONDUCTANCE MECHANOSENSITIVE CHANNEL | | Authors: | Wang, W, Dong, C, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Structure of an Open Form of an E. Coli Mechanosensitive Channel at 3.45 A Resolution.
Science, 321, 2008
|
|
1VJH
 
 | | Crystal structure of gene product of At1g24000 from Arabidopsis thaliana | | Descriptor: | Bet v I allergen family | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1H, 15N and 13C resonance assignments of the putative Bet v 1 family protein At1g24000.1 from Arabidopsis thaliana.
J.Biomol.Nmr, 32, 2005
|
|
2F2G
 
 | | X-Ray Structure of Gene Product From Arabidopsis Thaliana AT3G16990 | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SEED MATURATION PROTEIN PM36 HOMOLOG, SULFATE ION | | Authors: | Wesenberg, G.W, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-11-16 | | Release date: | 2005-12-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gene locus At3g16990 from Arabidopsis thaliana
Proteins, 57, 2004
|
|
