5IX8
 
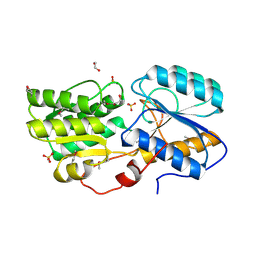 | | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822 | | Descriptor: | 1,2-ETHANEDIOL, Putative sugar ABC transport system, substrate-binding protein, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822
To Be Published
|
|
2AO9
 
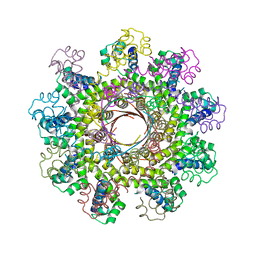 | |
8U01
 
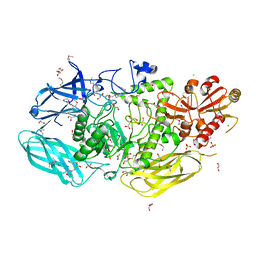 | | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain Containing Protein from Phocaeicola plebeius | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain
Containing Protein from Phocaeicola plebeius
To Be Published
|
|
4MPT
 
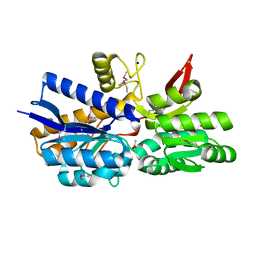 | | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I | | Descriptor: | ACETIC ACID, Putative leu/ile/val-binding protein, SODIUM ION | | Authors: | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I
To be Published
|
|
6W08
 
 | | Crystal Structure of Motility Associated Killing Factor E from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Joachimiak, G, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6B6L
 
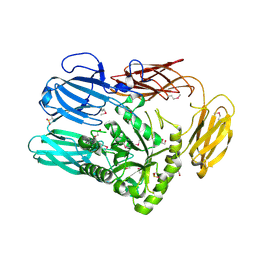 | | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Glycosyl hydrolase family 2, ... | | Authors: | Tan, K, Joachimiak, G, Nocek, B, Enddres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838
To Be Published
|
|
6NIO
 
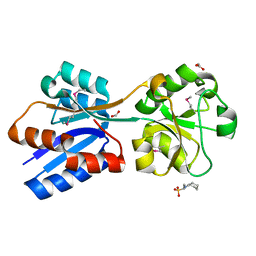 | | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, G, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis
To Be Published
|
|
2QM1
 
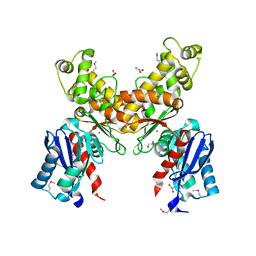 | | Crystal structure of glucokinase from Enterococcus faecalis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glucokinase, ... | | Authors: | Kim, Y, Joachimiak, G, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of Glucokinase from Enterococcus faecalis.
To be Published
|
|
2QQZ
 
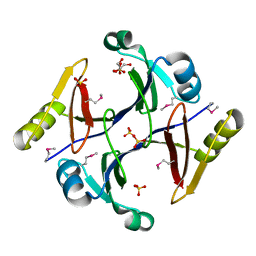 | | Crystal structure of putative glyoxalase family protein from Bacillus anthracis | | Descriptor: | GLYCEROL, Glyoxalase family protein, putative, ... | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Putative Glyoxalase Family Protein from Bacillus anthracis.
To be Published
|
|
5KZM
 
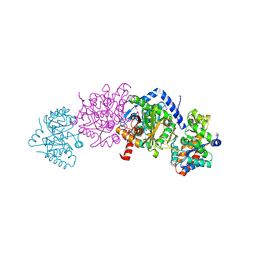 | | Crystal structure of Tryptophan synthase alpha-beta chain complex from Francisella tularensis | | Descriptor: | ACETATE ION, CALCIUM ION, Tryptophan synthase alpha chain, ... | | Authors: | Chang, C, Michalska, K, Joachimiak, G, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-10 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Conservation of the structure and function of bacterial tryptophan synthases.
Iucrj, 6, 2019
|
|
4XED
 
 | | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Peptidase M14, ... | | Authors: | Michalska, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20
To Be Published
|
|
2QQY
 
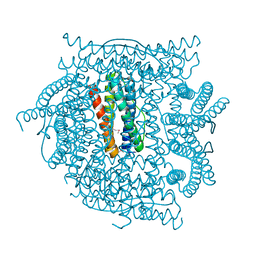 | | Crystal structure of ferritin like, diiron-carboxylate proteins from Bacillus anthracis str. Ames | | Descriptor: | Sigma B operon | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Ferritin like, Diiron-carboxylate Proteins from Bacillus anthracis str. Ames.
To be Published
|
|
1SQE
 
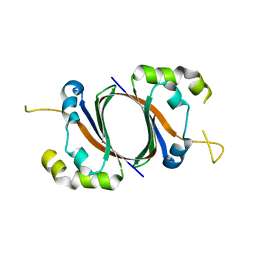 | | 1.5A Crystal Structure Of the protein PG130 from Staphylococcus aureus, Structural genomics | | Descriptor: | hypothetical protein PG130 | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases
J.Biol.Chem., 280, 2005
|
|
4J01
 
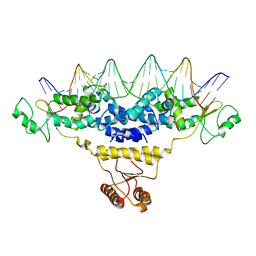 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 29mer DNA target | | Descriptor: | DNA (29-MER), SULFATE ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.246 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1R7L
 
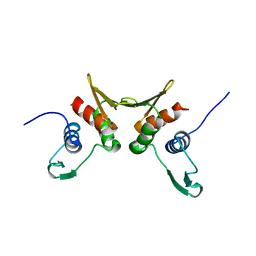 | | 2.0 A Crystal Structure of a Phage Protein from Bacillus cereus ATCC 14579 | | Descriptor: | Phage protein | | Authors: | Zhang, R, Joachimiak, G, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of phage protein BC1872 from Bacillus cereus, a singleton with new fold
Proteins, 64, 2006
|
|
3TY7
 
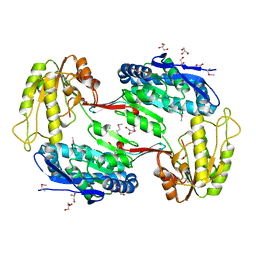 | | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Joachimiak, G, Jedrzejczak, R, Rubin, E, Ioerger, T, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus
To be Published
|
|
3B8F
 
 | | Crystal structure of the cytidine deaminase from Bacillus anthracis | | Descriptor: | Putative Blasticidin S deaminase | | Authors: | Zhang, R, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-01 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the cytidine deaminase from Bacillus anthracis.
To be Published
|
|
4KJM
 
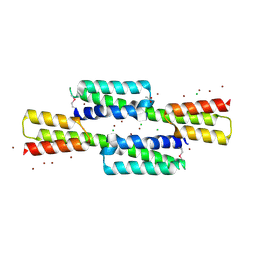 | | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB | | Descriptor: | ACETATE ION, CHLORIDE ION, Extracellular matrix-binding protein ebh, ... | | Authors: | Cymborowski, M, Shabalin, I.G, Joachimiak, G, Chruszcz, M, Gornicki, P, Zhang, R, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-03 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB
To be Published
|
|
5BMZ
 
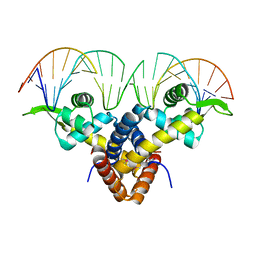 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA. | | Descriptor: | DNA (5'-D(P*GP*AP*AP*TP*AP*TP*CP*AP*GP*TP*TP*AP*AP*AP*CP*TP*GP*AP*TP*AP*TP*TP*C)-3'), HcaR protein | | Authors: | Kim, Y, Joachimiak, G, Biglow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA.
To Be Published
|
|
4IZZ
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 21mer DNA target | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*C)-3'), SULFATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J00
 
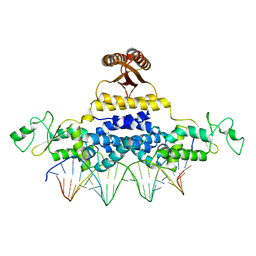 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 24mer DNA target | | Descriptor: | DNA (5'-D(*TP*GP*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*CP*C)-3'), MAGNESIUM ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KQC
 
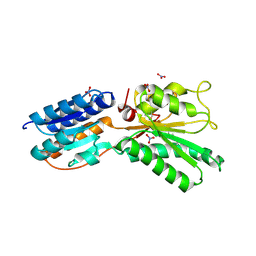 | | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii | | Descriptor: | NITRATE ION, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Osipiuk, J, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii
To be Published
|
|
1XBW
 
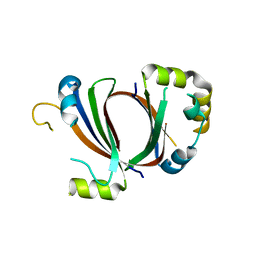 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | Descriptor: | hypothetical protein isdG | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
4XXT
 
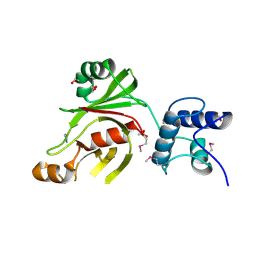 | | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from Clostridium acetobutylicum ATCC 824 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fusion of predicted Zn-dependent amidase/peptidase (Cell wall hydrolase/DD-carboxypeptidase family) and uncharacterized domain of ErfK family peptodoglycan-binding domain, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from from Clostridium acetobutylicum ATCC 824
To Be Published
|
|
4YE5
 
 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
