4KZT
 
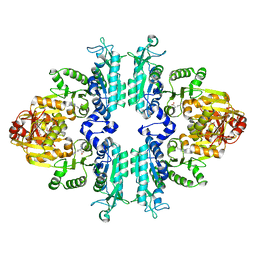 | | Structure mmNAGS bound with L-arginine | | Descriptor: | ARGININE, DI(HYDROXYETHYL)ETHER, N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2013-05-30 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of N-acetyl-L-glutamate synthase/kinase from Maricaulis maris with the allosteric inhibitor L-arginine bound.
Biochem.Biophys.Res.Commun., 437, 2013
|
|
3S6G
 
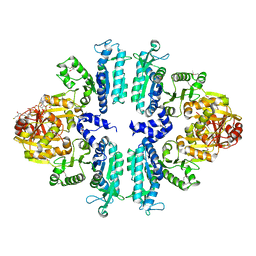 | | Crystal structures of Seleno-substituted mutant mmNAGS in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, MALONATE ION, ... | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6681 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6H
 
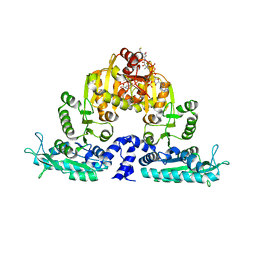 | | Crystal structure of native mmNAGS/k | | Descriptor: | COENZYME A, GLUTAMIC ACID, N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
2CSL
 
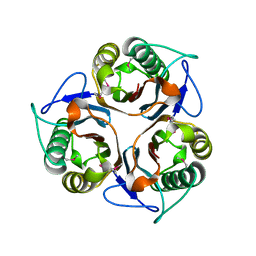 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | protein translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Jin, Z, Chrzas, J, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-22 | | Release date: | 2005-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
To be Published
|
|
4K30
 
 | | Structure of the N-acetyltransferase domain of human N-acetylglutamate synthase | | Descriptor: | N-ACETYL-L-GLUTAMATE, N-acetylglutamate synthase, mitochondrial | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of the N-acetyltransferase domain of human N-acetyl-L-glutamate synthase in complex with N-acetyl-L-glutamate provides insights into its catalytic and regulatory mechanisms.
Plos One, 8, 2013
|
|
6K6I
 
 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens
To Be Published
|
|
6K6J
 
 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion | | Descriptor: | BROMIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion
To Be Published
|
|
2CW9
 
 | | Crystal structure of human Tim44 C-terminal domain | | Descriptor: | PENTAETHYLENE GLYCOL, translocase of inner mitochondrial membrane | | Authors: | Handa, N, Kishishita, S, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Uchikubo, T, Takemoto, C, Jin, Z, Chrzas, J, Chen, L, Liu, Z.-J, Wang, B.-C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-17 | | Release date: | 2005-12-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human Tim44 C-terminal domain in complex with pentaethylene glycol: ligand-bound form.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6K6K
 
 | | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens
To Be Published
|
|
3S7Y
 
 | | Crystal structure of mmNAGS in Space Group P3121 at 4.3 A resolution | | Descriptor: | N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3077 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
4IWY
 
 | | SeMet-substituted RimK structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Ribosomal protein S6 modification protein, ... | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of Escherichia coli RimK, an ATP-grasp fold, l-glutamyl ligase enzyme.
Proteins, 81, 2013
|
|
4IWX
 
 | | Rimk structure at 2.85A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Ribosomal protein S6 modification protein, ... | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structure and function of Escherichia coli RimK, an ATP-grasp fold, l-glutamyl ligase enzyme.
Proteins, 81, 2013
|
|
7VCF
 
 | | Cryo-EM structure of Chlamydomonas TOC-TIC supercomplex | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, J, Yan, Z, Jin, Z, Zhang, Y. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes.
Cell, 185, 2022
|
|
3S6K
 
 | | Crystal structure of xcNAGS | | Descriptor: | Acetylglutamate kinase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.8018 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
6LZE
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11a | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhang, Y, Jing, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
7DRI
 
 | | Structure of SspE_CTD_41658 | | Descriptor: | DUF1524 domain | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRS
 
 | | Structure of SspE_40224 | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRR
 
 | | Structure of SspE-R100A protein | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
4GQ0
 
 | | Crystal structure of AKR1B10 complexed with NADP+ and Caffeic acid phenethyl ester | | Descriptor: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liping, Z, Xuehua, Z, Shangke, C, Jing, Z, Xiaopeng, H. | | Deposit date: | 2012-08-22 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of AKR1B10 complexed with NADP+ and Caffeic acid phenethyl ester
To be Published
|
|
3Q98
 
 | | Structure of ygeW encoded protein from E. coli | | Descriptor: | transcarbamylase | | Authors: | Li, Y, Jing, Z, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2011-01-07 | | Release date: | 2011-05-04 | | Last modified: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The ygeW encoded protein from Escherichia coli is a knotted ancestral catabolic transcarbamylase.
Proteins, 79, 2011
|
|
