1P1H
 
 | | Crystal structure of the 1L-myo-inositol/NAD+ complex | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jin, X, Geiger, J.H. | | Deposit date: | 2003-04-12 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P1J
 
 | | Crystal structure of the 1L-myo-inositol 1-phosphate synthase complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Inositol-3-phosphate synthase, ... | | Authors: | Jin, X, Geiger, J.H. | | Deposit date: | 2003-04-12 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P1I
 
 | |
1P1F
 
 | |
2ETA
 
 | |
5K6V
 
 | | Sidekick-1 immunoglobulin domains 1-4, crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein sidekick-1, ... | | Authors: | Jin, X, Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.208 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
5K6U
 
 | | Sidekick-1 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CESIUM ION, IODIDE ION, ... | | Authors: | Jin, X, Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
2ETB
 
 | | Crystal structure of the ankyrin repeat domain of TRPV2 | | Descriptor: | ACETIC ACID, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the N-terminal Ankyrin Repeat Domain of the TRPV2 Ion Channel.
J.Biol.Chem., 281, 2006
|
|
2IJR
 
 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | Descriptor: | Hypothetical protein api92 | | Authors: | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
2ETC
 
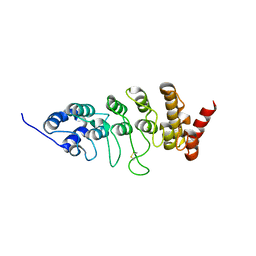 | | Crystal structure of the ankyrin repeat domain of TRPV2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the N-terminal Ankyrin Repeat Domain of the TRPV2 Ion Channel.
J.Biol.Chem., 281, 2006
|
|
4NUP
 
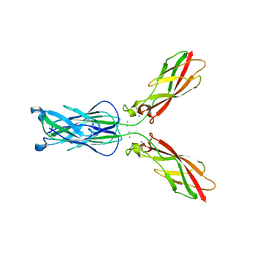 | |
4NUM
 
 | | Crystal structure of mouse N-cadherin EC1-2 A78SI92M | | Descriptor: | CALCIUM ION, Cadherin-2 | | Authors: | Jin, X. | | Deposit date: | 2013-12-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and energetic determinants of adhesive binding specificity in type I cadherins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NUQ
 
 | | Crystal structure of mouse N-cadherin EC1-2 W2F | | Descriptor: | CALCIUM ION, Cadherin-2 | | Authors: | Jin, X. | | Deposit date: | 2013-12-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Structural and energetic determinants of adhesive binding specificity in type I cadherins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3Q2V
 
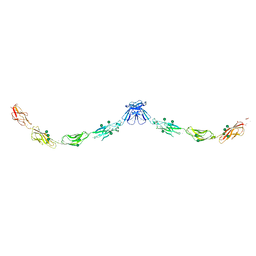 | | Crystal structure of mouse E-cadherin ectodomain | | Descriptor: | CALCIUM ION, Cadherin-1, MANGANESE (II) ION, ... | | Authors: | Jin, X, Harrison, O.J, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
3Q2W
 
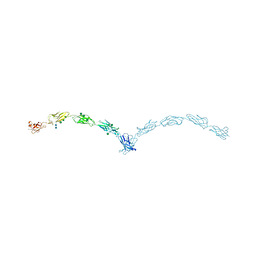 | | Crystal structure of mouse N-cadherin ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
3QRB
 
 | | crystal structure of E-cadherin EC1-2 P5A P6A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-1, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2011-02-17 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular design principles underlying beta-strand swapping in the adhesive dimerization of cadherins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2NPO
 
 | | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Acetyltransferase | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
2NLY
 
 | | Crystal structure of protein BH1492 from Bacillus halodurans, Pfam DUF610 | | Descriptor: | Divergent polysaccharide deacetylase hypothetical protein, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-20 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein BH1492 from Bacillus halodurans C-125
To be Published
|
|
2NS9
 
 | | Crystal structure of protein APE2225 from Aeropyrum pernix K1, Pfam COXG | | Descriptor: | Hypothetical protein APE2225, PHOSPHATE ION | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein APE2225 from Aeropyrum pernix K1
To be Published
|
|
2NYJ
 
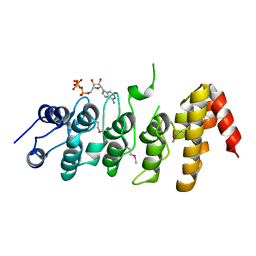 | | Crystal structure of the ankyrin repeat domain of TRPV1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Ankyrin Repeats of TRPV1 Bind Multiple Ligands and Modulate Channel Sensitivity.
Neuron, 54, 2007
|
|
2OCE
 
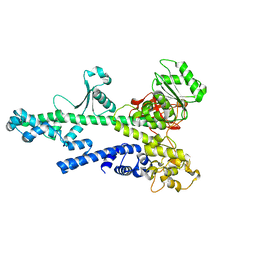 | |
7YE7
 
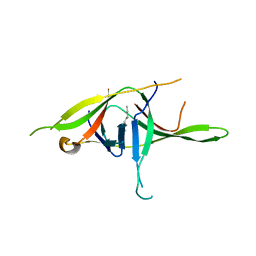 | | Crystal structure of SARS-CoV-2 soluble dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein, nonane | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
7YE8
 
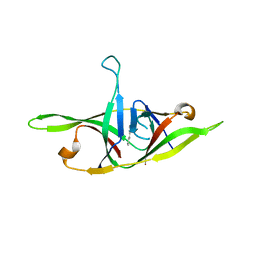 | | Crystal structure of SARS-CoV-2 refolded dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
2HY3
 
 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I0O
 
 | | Crystal structure of Anopheles gambiae Ser/Thr phosphatase complexed with Zn2+ | | Descriptor: | Ser/Thr phosphatase, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
