1UGW
 
 | | Crystal structure of jacalin- Gal complex | | Descriptor: | Agglutinin alpha chain, Agglutinin alpha-chain, Agglutinin beta-3 chain, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1UH1
 
 | | Crystal structure of jacalin- GalNAc-beta(1-3)-Gal-alpha-O-Me complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-methyl alpha-D-galactopyranoside, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1UH0
 
 | | Crystal structure of jacalin- Me-alpha-GalNAc complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, methyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1UGY
 
 | | Crystal structure of jacalin- mellibiose (Gal-alpha(1-6)-Glc) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
8S30
 
 | |
8S31
 
 | |
8QSU
 
 | |
8QSW
 
 | |
8QSV
 
 | |
1M26
 
 | | Crystal structure of jacalin-T-antigen complex | | Descriptor: | Jacalin, alpha chain, beta chain, ... | | Authors: | Jeyaprakash, A.A, Rani, P.G, Reddy, G.B, Banumathi, S, Betzel, C, Surolia, A, Vijayan, M. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the jacalin-T-antigen complex and a
comparative study of lectin-T-antigen complexs
J.Mol.Biol., 321, 2002
|
|
1WS4
 
 | | Crystal structure of Jacalin- Me-alpha-Mannose complex: Promiscuity vs Specificity | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, methyl alpha-D-galactopyranoside, ... | | Authors: | Jeyaprakash, A.A, Jayashree, G, Mahanta, S.K, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2004-10-31 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the energetics of jacalin-sugar interactions: promiscuity versus specificity
J.Mol.Biol., 347, 2005
|
|
1WS5
 
 | | Crystal structure of Jacalin-Me-alpha-Mannose complex: Promiscuity vs Specificity | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, methyl alpha-D-mannopyranoside | | Authors: | Jeyaprakash, A.A, Jayashree, G, Mahanta, S.K, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2004-10-31 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the energetics of jacalin-sugar interactions: promiscuity versus specificity
J.Mol.Biol., 347, 2005
|
|
1VBP
 
 | | Crystal structure of artocarpin-mannopentose complex | | Descriptor: | SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Jeyaprakash, A.A, Srivastav, A, Surolia, A, Vijayan, M. | | Deposit date: | 2004-02-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity
J.Mol.Biol., 338, 2004
|
|
1VBO
 
 | | Crystal structure of artocarpin-mannotriose complex | | Descriptor: | alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, artocarpin | | Authors: | Jeyaprakash, A.A, Srivastav, A, Surolia, A, Vijayan, M. | | Deposit date: | 2004-02-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity
J.Mol.Biol., 338, 2004
|
|
1UGX
 
 | | Crystal structure of jacalin- Me-alpha-T-antigen (Gal-beta(1-3)-GalNAc-alpha-o-Me) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
2QFA
 
 | | Crystal structure of a Survivin-Borealin-INCENP core complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Baculoviral IAP repeat-containing protein 5, Borealin, ... | | Authors: | Jeyaprakash, A.A, Klein, U.R, Lindner, D, Ebert, J, Nigg, E.A, Conti, E. | | Deposit date: | 2007-06-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a Survivin-Borealin-INCENP Core Complex Reveals How Chromosomal Passengers Travel Together.
Cell(Cambridge,Mass.), 131, 2007
|
|
6XWU
 
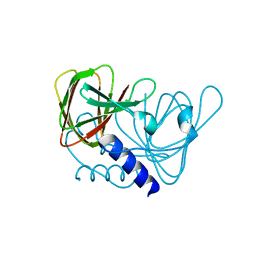 | | Crystal structure of drosophila melanogaster CENP-C cumin domain | | Descriptor: | RE68959p | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
6XWS
 
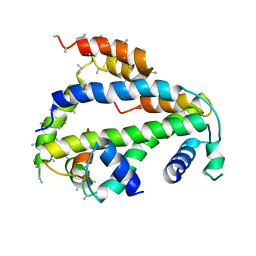 | | Crystal Structure of Drosophila CAL1 1-160 bound to CENP-A/H4 | | Descriptor: | Chromosome alignment defect 1,Chromosome alignment defect 1, Histone H3-like centromeric protein cid, Histone H4 | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
6XWV
 
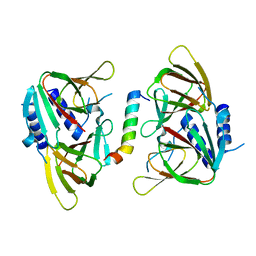 | | Crystal structure of drosophila melanogaster CENP-C bound to CAL1 | | Descriptor: | Calmodulin, Ryanodine Receptor 2 | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
6XWT
 
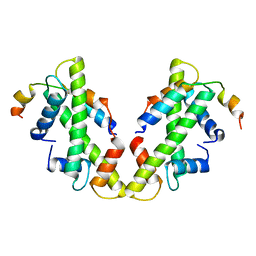 | | drosophila melanogaster CENP-A/H4 bound to N-terminal CAL1 fragment | | Descriptor: | Chromosome alignment defect 1, Histone H3-like centromeric protein cid, Histone H4 | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
4AJ5
 
 | | Crystal structure of the Ska core complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 2, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 3 | | Authors: | Jeyaprakash, A.A, Santamaria, A, Jayachandran, U, Chan, Y.W, Benda, C, Nigg, E.A, Conti, E. | | Deposit date: | 2012-02-15 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural and Functional Organization of the Ska Complex, a Key Component of the Kinetochore-Microtubule Interface.
Mol.Cell, 46, 2012
|
|
4A0J
 
 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A0I
 
 | | Crystal structure of Survivin bound to the N-terminal tail of hSgo1 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, SHUGOSHIN-LIKE 1, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A0N
 
 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4HR6
 
 | | Crystal structure of snake gourd (Trichosanthes anguina) seed lectin, a three chain homologue of type II RIPs | | Descriptor: | LECTIN, methyl alpha-D-galactopyranoside | | Authors: | Sharma, A, Pohlentz, G, Bobbili, K.B, Jeyaprakash, A.A, Chandran, T, Mormann, M, Swamy, M.J, Vijayan, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The sequence and structure of snake gourd (Trichosanthes anguina) seed lectin, a three-chain nontoxic homologue of type II RIPs.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
