6CFZ
 
 | |
4V58
 
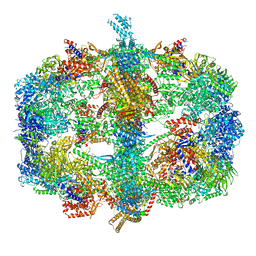 | | Crystal structure of fatty acid synthase from thermomyces lanuginosus at 3.1 angstrom resolution. | | 分子名称: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE | | 著者 | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | 登録日 | 2007-03-09 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
4V59
 
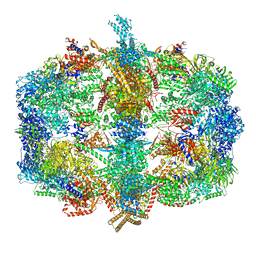 | | Crystal structure of fatty acid synthase complexed with nadp+ from thermomyces lanuginosus at 3.1 angstrom resolution. | | 分子名称: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | 登録日 | 2007-03-09 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6U1X
 
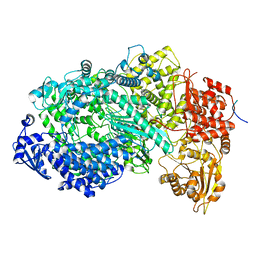 | | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | | 分子名称: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Jenni, S, Bloyet, L.M, Dias-Avalos, R, Liang, B, Wheelman, S.P.J, Grigorieff, N, Harrison, S.C. | | 登録日 | 2019-08-17 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Cell Rep, 30, 2020
|
|
5AOQ
 
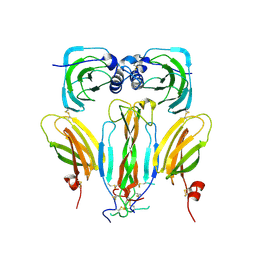 | | Structural basis of neurohormone perception by the receptor tyrosine kinase Torso | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PREPROPTTH, TORSO | | 著者 | Jenni, S, Goyal, Y, von Grotthuss, M, Shvartsman, S.Y, Klein, D.E. | | 登録日 | 2015-09-11 | | 公開日 | 2015-11-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis of Neurohormone Perception by the Receptor Tyrosine Kinase Torso.
Mol.Cell, 60, 2015
|
|
7UMK
 
 | | Structure of vesicular stomatitis virus (helical reconstruction, 4.1 A resolution) | | 分子名称: | Matrix protein, Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | 著者 | Jenni, S, Horwitz, J.A, Bloyet, L.-M, Whelan, S.P.J, Harrison, S.C. | | 登録日 | 2022-04-07 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Nat Commun, 13, 2022
|
|
7UML
 
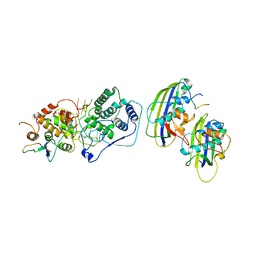 | | Structure of vesicular stomatitis virus (local reconstruction, 3.5 A resolution) | | 分子名称: | Matrix protein, Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | 著者 | Jenni, S, Horwitz, J.A, Bloyet, L.-M, Whelan, S.P.J, Harrison, S.C. | | 登録日 | 2022-04-07 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Nat Commun, 13, 2022
|
|
2CDH
 
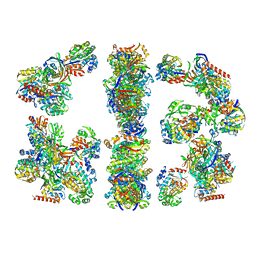 | | ARCHITECTURE OF THE THERMOMYCES LANUGINOSUS FUNGAL FATTY ACID SYNTHASE AT 5 ANGSTROM RESOLUTION. | | 分子名称: | DEHYDRATASE, ENOYL REDUCTASE, KETOACYL REDUCTASE, ... | | 著者 | Jenni, S, Leibundgut, M, Maier, T, Ban, N. | | 登録日 | 2006-01-24 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Architecture of a Fungal Fatty Acid Synthase at 5 A Resolution.
Science, 311, 2006
|
|
7UMS
 
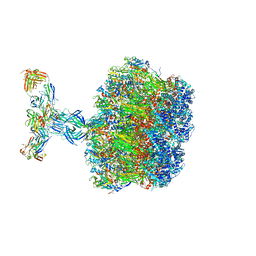 | | Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 in complex with antibody 41 - Upright conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fab 41 heavy chain, ... | | 著者 | Jenni, S, Zongli, L, Wang, Y, Bessey, T, Salgado, E.N, Schmidt, A.G, Greenberg, H.B, Jiang, B, Harrison, S.C. | | 登録日 | 2022-04-07 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.
J.Virol., 96, 2022
|
|
7UMT
 
 | | Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 - Reversed conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | 著者 | Jenni, S, Zongli, L, Wang, Y, Bessey, T, Salgado, E.N, Schmidt, A.G, Greenberg, H.B, Jiang, B, Harrison, S.C. | | 登録日 | 2022-04-07 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.
J.Virol., 96, 2022
|
|
5UK1
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
3ZVR
 
 | |
6PP7
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6POD
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-03 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP5
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PO3
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-03 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PPE
 
 | | ClpP and ClpX IGF loop in ClpX-ClpP complex with D7 symmetry | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-06 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6POS
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PO1
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-03 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP8
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP6
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
