5JOD
 
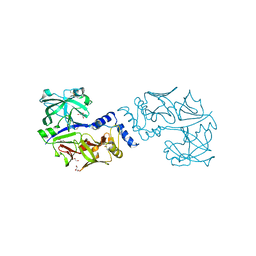 | | Structure of proplasmepsin IV from Plasmodium falciparum | | Descriptor: | GLYCEROL, Proplasmepsin IV | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5JXV
 
 | | Solid-state MAS NMR structure of immunoglobulin beta 1 binding domain of protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Andreas, L.B, Jaudzems, K, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2KZF
 
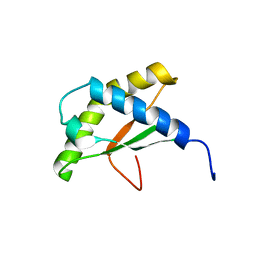 | | Solution NMR structure of the thermotoga maritima protein TM0855 a putative ribosome binding factor A | | Descriptor: | Ribosome-binding factor A | | Authors: | Serrano, P, Jaudzems, K, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the thermotoga maritima protein TM0855
To be Published
|
|
2KYS
 
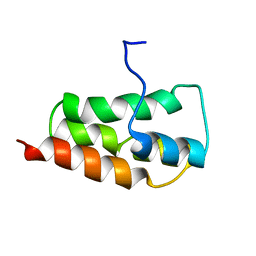 | | NMR Structure of the SARS Coronavirus Nonstructural Protein Nsp7 in Solution at pH 6.5 | | Descriptor: | Non-structural protein 7 | | Authors: | Johnson, M.A, Jaudzems, K, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the SARS-CoV Nonstructural Protein 7 in Solution at pH 6.5.
J.Mol.Biol., 402, 2010
|
|
2KTS
 
 | | NMR structure of the protein NP_415897.1 | | Descriptor: | Heat shock protein hslJ | | Authors: | Serrano, P, Jaudzems, K, Geralt, M, Horst, R, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-02-06 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_415897.1
To be Published
|
|
2LTH
 
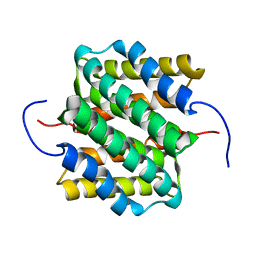 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 5.5 | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Otikovs, M, Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S, Johansson, J. | | Deposit date: | 2012-05-25 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation.
Nat Commun, 5, 2014
|
|
2MX9
 
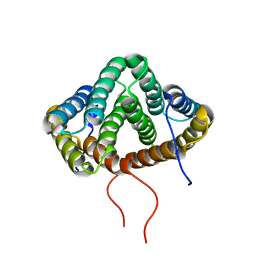 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 5.5 | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-19 | | Method: | SOLUTION NMR | | Cite: | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
2MX8
 
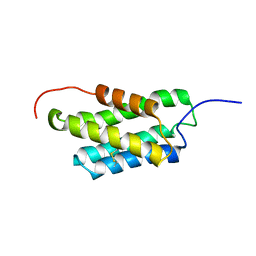 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 7.2 | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
2MFZ
 
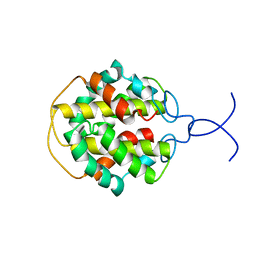 | | NMR structure of C-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Andersson, M, Chen, G, Landreh, M, Nordling, K, Kronqvist, N, Westermark, P, Jornvall, H, Knight, S, Ridderstrale, Y, Holm, L, Meng, Q, Chesler, M, Johansson, J, Rising, A. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains
Plos Biol., 12, 2014
|
|
2K7R
 
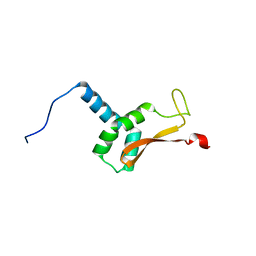 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
