2JTA
 
 | |
5JDP
 
 | | E73V mutant of the human voltage-dependent anion channel | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Jaremko, M, Jaremko, L, Villinger, S, Schmidt, C, Giller, K, Griesinger, C, Becker, S, Zweckstetter, M. | | Deposit date: | 2016-04-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Determination of the Dynamic Structure of Membrane Proteins.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5JDX
 
 | | PigG holo | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Putative peptidyl carrier protein | | Authors: | Jaremko, M.J, Lee, D.J, Burkart, M.D. | | Deposit date: | 2016-04-17 | | Release date: | 2017-08-23 | | Last modified: | 2019-12-04 | | Method: | SOLUTION NMR | | Cite: | PigG holo
To Be Published
|
|
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
2LYQ
 
 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYR
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYP
 
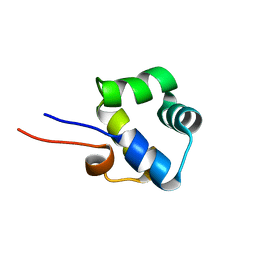 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
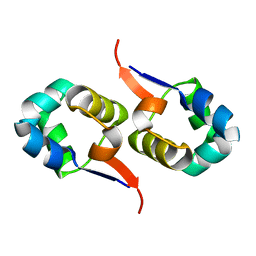 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
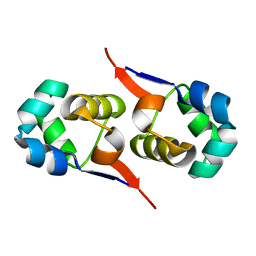 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2N02
 
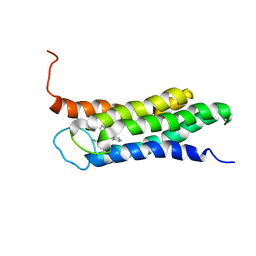 | | Solution structure of the A147T variant of the mitochondrial translocator protein (tspo) in complex with pk11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Integrity of the A147T Polymorph of Mammalian TSPO.
Chembiochem, 16, 2015
|
|
2LYL
 
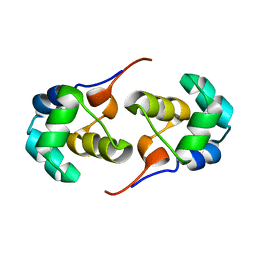 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
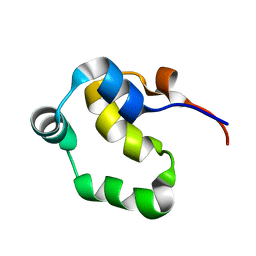 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2MGY
 
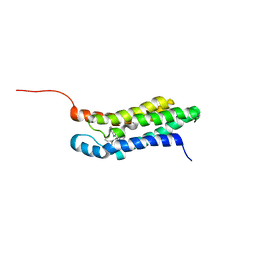 | | Solution structure of the mitochondrial translocator protein (TSPO) in complex with its high-affinity ligand PK11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the mitochondrial translocator protein in complex with a diagnostic ligand.
Science, 343, 2014
|
|
7JPS
 
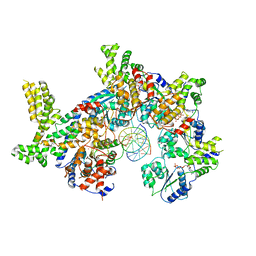 | |
7JPQ
 
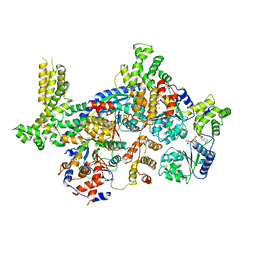 | | ORC-O2-5: Human Origin Recognition Complex (ORC) with subunits 2,3,4,5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 2, ... | | Authors: | Jaremko, M.J, Joshua-Tor, L. | | Deposit date: | 2020-08-09 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The dynamic nature of the human Origin Recognition Complex revealed through five cryoEM structures.
Elife, 9, 2020
|
|
7JPO
 
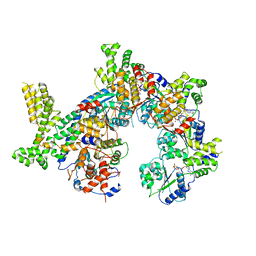 | |
7JPP
 
 | |
7JPR
 
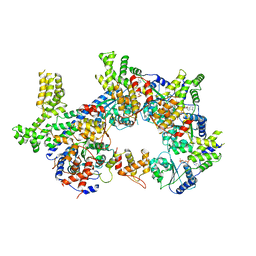 | |
2N5I
 
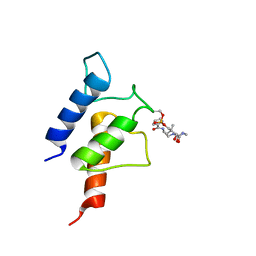 | | PltL-pyrrolyl | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(1H-pyrrol-2-ylcarbonyl)amino]ethyl}amino)propyl]amino}butyl dihydrogen phosphate, Peptidyl carrier protein PltL | | Authors: | Jaremko, M.J, Lee, D.J, Burkart, M.D. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate Sequestration in the Pyoluteorin Type II Peptidyl Carrier Protein PltL.
J.Am.Chem.Soc., 137, 2015
|
|
2N5H
 
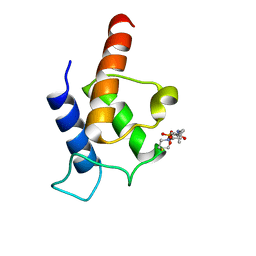 | | PltL-holo | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Peptidyl carrier protein PltL | | Authors: | Jaremko, M.J, Lee, D.J, Burkart, M.D. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate Sequestration in the Pyoluteorin Type II Peptidyl Carrier Protein PltL.
J.Am.Chem.Soc., 137, 2015
|
|
2V31
 
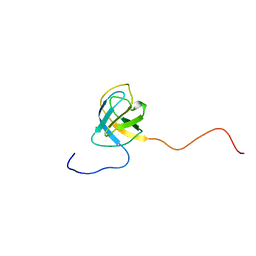 | | Structure of First Catalytic Cysteine Half-domain of mouse ubiquitin- activating enzyme | | Descriptor: | UBIQUITIN-ACTIVATING ENZYME E1 X | | Authors: | Jaremko, L, Jaremko, M, Wojciechowski, W, Filipek, R, Szczepanowski, R.H, Bochtler, M, Zhukov, I. | | Deposit date: | 2007-06-11 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of First Catalytic Cysteine Half-Domain of Mouse Ubiquitin-Activating Enzyme
To be Published
|
|
2RQS
 
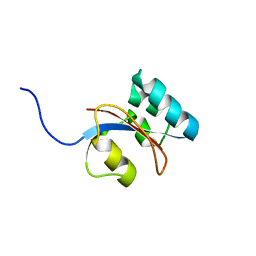 | | 3D structure of Pin from the psychrophilic archeon Cenarcheaum symbiosum (CsPin) | | Descriptor: | Parvulin-like peptidyl-prolyl isomerase | | Authors: | Zhukov, I, Jaremko, L, Jaremko, M, Mueller, J.W, Bayer, P. | | Deposit date: | 2009-11-17 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the First Archaeal Parvulin Reveal a New Functionally Important Loop in Parvulin-type Prolyl Isomerases
J.Biol.Chem., 286, 2011
|
|
2JT9
 
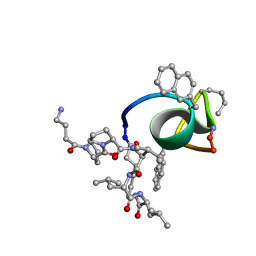 | | NMR structure of immunosuppressory peptide containing cyclolinopeptide X and antennapedia(43-58) sequences | | Descriptor: | 5-mer immunosuppressory peptide from cyclolinopeptide X, 6-AMINOHEXANOIC ACID, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Jaremko, L, Jaremko, M, Zhukov, I, Cebrat, M. | | Deposit date: | 2007-07-21 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of immunosuppressory peptide containing cyclolinopeptide X
and antennapedia(43-58) sequences
To be Published
|
|
2LLS
 
 | | solution structure of human apo-S100A1 C85M | | Descriptor: | Protein S100-A1 | | Authors: | Budzinska, M, Jaremko, L, Jaremko, M, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical Shift Assignments and solution structure of human apo-S100A1 C85M mutant
To be Published
|
|
2M1I
 
 | |
