2ZD8
 
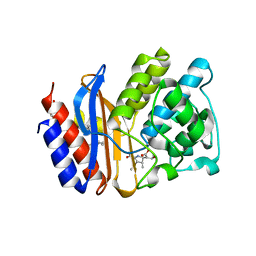 | | SHV-1 class A beta-lactamase complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Nukaga, M, Knox, J.R, Hujer, A, Bonomo, R.A. | | Deposit date: | 2007-11-20 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Inhibition of class A beta-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1.
J.Am.Chem.Soc., 130, 2008
|
|
6OAB
 
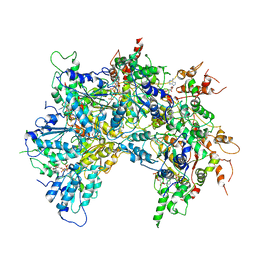 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OHI
 
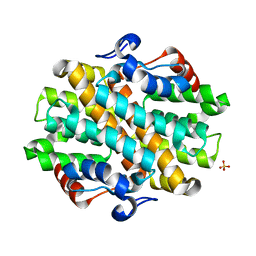 | | Crystal Structure of the Debrominase Bmp8 (Apo) | | Descriptor: | Debrominase Bmp8, SULFATE ION | | Authors: | Chekan, J.R, Moore, B.S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Bacterial Tetrabromopyrrole Debrominase Shares a Reductive Dehalogenation Strategy with Human Thyroid Deiodinase.
Biochemistry, 58, 2019
|
|
4B9H
 
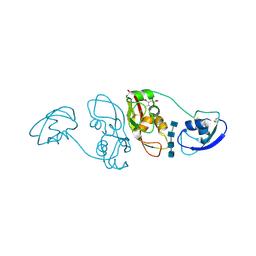 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer: I3C heavy atom derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
2XL3
 
 | | WDR5 IN COMPLEX WITH AN RBBP5 PEPTIDE AND HISTONE H3 PEPTIDE | | Descriptor: | GLYCEROL, HISTONE H3.1, RETINOBLASTOMA-BINDING PROTEIN 5, ... | | Authors: | Odho, Z, Southall, S.M, Wilson, J.R. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterisation of a Novel Wdr5 Binding Site that Recruits Rbbp5 Through a Conserved Motif and Enhances Methylation of H3K4 by Mll1.
J.Biol.Chem., 285, 2010
|
|
2X63
 
 | | Crystal structure of the sialyltransferase CST-II N51A in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
2X62
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE CST-II Y81F IN COMPLEX WITH CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, N3-PROTONATED CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
6F0P
 
 | | Botulinum neurotoxin A4 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Davies, J.R, Rees, J, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
2WYJ
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1S)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2WUV
 
 | | Crystallographic analysis of counter-ion effects on subtilisin enzymatic action in acetonitrile | | Descriptor: | ACETONITRILE, CALCIUM ION, CESIUM ION, ... | | Authors: | Cianci, M, Tomaszewki, B, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystallographic Analysis of Counterion Effects on Subtilisin Enzymatic Action in Acetonitrile.
J.Am.Chem.Soc., 132, 2010
|
|
2WUW
 
 | | Crystallographic analysis of counter-ion effects on subtilisin enzymatic action in acetonitrile (native data) | | Descriptor: | ACETONITRILE, CALCIUM ION, SODIUM ION, ... | | Authors: | Cianci, M, Tomaszewki, B, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystallographic Analysis of Counterion Effects on Subtilisin Enzymatic Action in Acetonitrile.
J.Am.Chem.Soc., 132, 2010
|
|
4BUP
 
 | | A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases | | Descriptor: | GLYCEROL, HISTONE-LYSINE N-METHYLTRANSFERASE SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Southall, S.M, Cronin, N.B, Wilson, J.R. | | Deposit date: | 2013-06-21 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.166 Å) | | Cite: | A Novel Route to Product Specificity in the Suv4-20 Family of Histone H4K20 Methyltransferases.
Nucleic Acids Res., 42, 2014
|
|
2XVR
 
 | | Phage T7 empty mature head shell | | Descriptor: | MAJOR CAPSID PROTEIN 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Molecular Rearrangements Involved in the Capsid Shell Maturation of Bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
6F0O
 
 | | Botulinum neurotoxin A3 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, Bontoxilysin A, PENTAETHYLENE GLYCOL, ... | | Authors: | Davies, J.R, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
2XTH
 
 | | K2PtBr6 binding to lysozyme | | Descriptor: | HEXABROMOPLATINATE(IV), LYSOZYME C | | Authors: | Helliwell, J.R, Bell, A.M.T, Bryant, P, Fisher, S, Habash, J, Helliwell, M, Margiolaki, I, Kaenket, S, Watier, Y, Wright, J, Yalamanchili, S.K. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-Dependent Analysis of K2Ptbr6 Binding to Lysozyme Studied by Protein Powder and Single Crystal X-Ray Analysis
Z.Kristallogr., 225, 2010
|
|
2ROY
 
 | | TRANSTHYRETIN (ALSO CALLED PREALBUMIN) COMPLEX WITH 3',5'-DINITRO-N-ACETYL-L-THYRONINE | | Descriptor: | 3',5'-DINITRO-N-ACETYL-L-THYRONINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 1996-10-23 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of human transthyretin complexed with thyroxine at 2.0 A resolution and 3',5'-dinitro-N-acetyl-L-thyronine at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6BH3
 
 | |
4B5Z
 
 | | Structure of the apo-rFnBPA(189-505) from Staphylococcus aureus | | Descriptor: | FIBRONECTIN-BINDING PROTEIN A | | Authors: | Stemberk, V, Moroz, O, Atkin, K.E, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2012-08-08 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evidence for Steric Regulation of Fibrinogen Binding to Staphylococcus Aureus Fibronectin-Binding Protein a (Fnbpa).
J.Biol.Chem., 289, 2014
|
|
6BGW
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(4,4-difluoropiperidin-1-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N41) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(4,4-difluoropiperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGV
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N40) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH5
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(3-(piperidin-1-yl)propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N48) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[3-(piperidin-1-yl)propoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6P64
 
 | |
2WNF
 
 | | Crystal Structure of a Mammalian Sialyltransferase in complex with Gal-beta-1-3GalNAc-ortho-nitrophenol | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, HYDROXY(2-HYDROXYPHENYL)OXOAMMONIUM, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2WML
 
 | | Crystal Structure of a Mammalian Sialyltransferase | | Descriptor: | CHLORIDE ION, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE -ALPHA-2,3-SIALYLTRANSFERASE, GLYCEROL | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-01 | | Release date: | 2009-10-13 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6BGY
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound 46) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(S)-(2-chlorophenyl){2-[(2R)-1-methylpyrrolidin-2-yl]ethoxy}methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
