3K15
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K05
 
 | |
3HZZ
 
 | |
3IR8
 
 | | Red fluorescent protein mKeima at pH 7.0 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Henderson, J.N, Osborn, M.F, Koon, N, Gepshtein, R, Huppert, D, Remington, S.J. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited state proton transfer in the red fluorescent protein mKeima.
J.Am.Chem.Soc., 131, 2009
|
|
3HZX
 
 | |
2AKJ
 
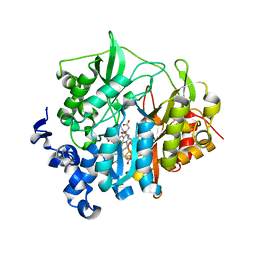 | | Structure of spinach nitrite reductase | | Descriptor: | Ferredoxin--nitrite reductase, chloroplast, IRON/SULFUR CLUSTER, ... | | Authors: | Swamy, U, Wang, M, Tripathy, J.N, Kim, S.-K, Hirasawa, M, Knaff, D.B, Allen, J.P. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Spinach Nitrite Reductase: Implications for Multi-electron Reactions by the Iron-Sulfur:Siroheme Cofactor
Biochemistry, 44, 2005
|
|
2BER
 
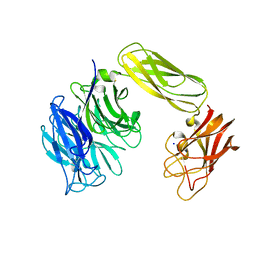 | | Y370G Active Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). | | Descriptor: | BACTERIAL SIALIDASE, N-acetyl-beta-neuraminic acid, SODIUM ION | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G.L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of Action of an Inverting Mutant Sialidase.
Biochemistry, 44, 2005
|
|
5AF4
 
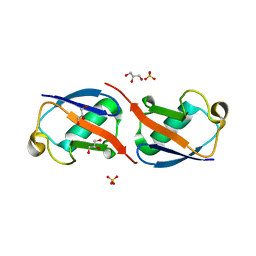 | | Structure of Lys33-linked diUb | | Descriptor: | GLYCEROL, SULFATE ION, UBIQUITIN | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
9FFH
 
 | | Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography.
Biorxiv, 2024
|
|
9FFG
 
 | | Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Byrom, L, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography.
Biorxiv, 2024
|
|
7O7G
 
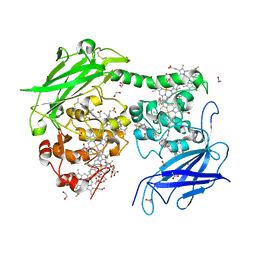 | | Crystal structure of the Shewanella oneidensis MR1 MtrC mutant H561M | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Edwards, M.J, van Wonderen, J.H, Newton-Payne, S.E, Butt, J.N, Clarke, T.A. | | Deposit date: | 2021-04-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanosecond heme-to-heme electron transfer rates in a multiheme cytochrome nanowire reported by a spectrally unique His/Met-ligated heme.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NTF
 
 | | Cryo-EM structure of unliganded O-GlcNAc transferase | | Descriptor: | Isoform 1 of UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Meek, R.W, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.32 Å) | | Cite: | Cryo-EM structure provides insights into the dimer arrangement of the O-linked beta-N-acetylglucosamine transferase OGT.
Nat Commun, 12, 2021
|
|
3F8L
 
 | | Crystal Structure of the Effector Domain of PhnF from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, HTH-type transcriptional repressor phnF, SULFATE ION | | Authors: | Busby, J.N, Gebhard, S, Cook, G.M, Baker, E.N, Lott, S.J, Money, V.A. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of PhnF, a GntR-family transcription regulator in Mycobacterium smegmatis
To be Published
|
|
3FS0
 
 | |
3FTM
 
 | |
7P92
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain up | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P8N
 
 | | TmHydABC- T. maritima hydrogenase with bridge closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P91
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P5H
 
 | | TmHydABC- D2 map | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-14 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7OVM
 
 | | Protein kinase MKK7 in complex with cyclobutyl-substituted indazole | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[(1-cyclobutyl-1,2,3-triazol-4-yl)methyl]-3-(1~{H}-indazol-3-yl)-5-(propanoylamino)benzamide | | Authors: | Buehrmann, M, Wiese, J.N, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7OVN
 
 | | Protein kinase MKK7 in complex with tolyl-substituted indazole | | Descriptor: | 3-(1~{H}-indazol-3-yl)-~{N}-[[1-(2-methylphenyl)-1,2,3-triazol-4-yl]methyl]-5-(propanoylamino)benzamide, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Buehrmann, M, Wiese, J.N, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7OVJ
 
 | | Protein kinase MKK7 in complex with difluoro-phenethyltriazole-substituted pyrazolopyrimidine | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-[1-[2,2-bis(fluoranyl)-2-phenyl-ethyl]-1,2,3-triazol-4-yl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7OVL
 
 | | Protein kinase MKK7 in complex with methoxycyclohexyl-substituted indazole | | Descriptor: | 3-(2~{H}-indazol-3-yl)-~{N}-[[1-[(1~{R},2~{R})-2-methoxycyclohexyl]-1,2,3-triazol-4-yl]methyl]-5-(propanoylamino)benzamide, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Buehrmann, M, Wiese, J.N, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7P4R
 
 | | Ultra High Resolution X-ray Structure of Orthorhombic Bovine Pancreatic Ribonuclease at 100K | | Descriptor: | ETHANOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Lisgarten, D.R, Palmer, R.A, Cooper, J.B, Naylor, C.E, Howlin, B.J, Lisgarten, J.N, Najmudin, S, Lobley, C.M.C. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultra-high resolution X-ray structure of orthorhombic bovine pancreatic Ribonuclease A at 100K.
BMC Chem, 17, 2023
|
|
5AF5
 
 | | Structure of Lys33-linked triUb S.G. P 212121 | | Descriptor: | POLYUBIQUITIN-C | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
