5DSE
 
 | | Crystal Structure of the TTC7B/Hyccin Complex | | Descriptor: | Hyccin, Tetratricopeptide repeat protein 7B | | Authors: | Wu, X, Baskin, J.M, Reinisch, K.M, De Camilli, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane.
Nat.Cell Biol., 18, 2016
|
|
4XBU
 
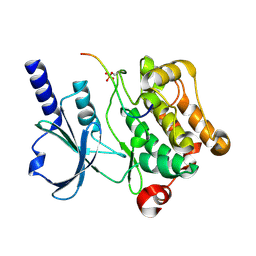 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | Descriptor: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4C6Y
 
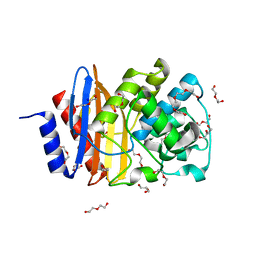 | | Ancestral PNCA (last common ancestors of Gram-positive and Gram- negative bacteria) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
4XGC
 
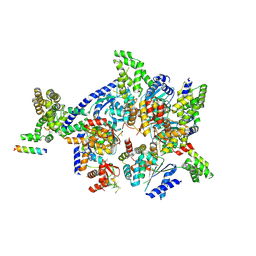 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
4C75
 
 | | Consensus (ALL-CON) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
4XBR
 
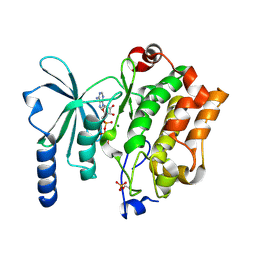 | | In cellulo Crystal Structure of PAK4 in complex with Inka | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein FAM212A,Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XDS
 
 | | Deoxyguanosinetriphosphate Triphosphohydrolase from Escherichia coli with Nickel | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, NICKEL (II) ION, SULFATE ION | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
4XTD
 
 | | Structure of the covalent intermediate E-XMP* of the IMP dehydrogenase of Ashbya gossypii | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-01-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XKT
 
 | | E coli BFR variant Y149F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Bradley, J.M, Hemmings, A.M, Le Brun, N.E. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Three Aromatic Residues are Required for Electron Transfer during Iron Mineralization in Bacterioferritin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XY3
 
 | | Structure of ESX-1 secreted protein EspB | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Wagner, J.M, Korotkov, K.V. | | Deposit date: | 2015-02-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of EspB, a secreted substrate of the ESX-1 secretion system of Mycobacterium tuberculosis.
J.Struct.Biol., 191, 2015
|
|
4XTI
 
 | | Structure of IMP dehydrogenase of Ashbya gossypii with IMP bound to the active site | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, POTASSIUM ION | | Authors: | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-01-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XKU
 
 | | E coli BFR variant Y114F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Hemmings, A.M, Le Brun, N.E, Bradley, J.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three Aromatic Residues are Required for Electron Transfer during Iron Mineralization in Bacterioferritin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XUC
 
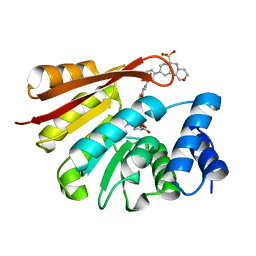 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd18 (1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one) | | Descriptor: | 1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4XVX
 
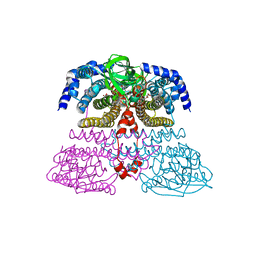 | | Crystal structure of an acyl-ACP dehydrogenase | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acyl-[acyl-carrier-protein] dehydrogenase MbtN, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Chai, A, Johnston, J.M, Bunker, R.D, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A covalent adduct of MbtN, an acyl-ACP dehydrogenase from Mycobacterium tuberculosis, reveals an unusual acyl-binding pocket.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4D89
 
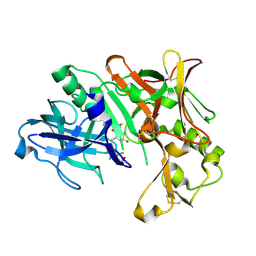 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BXD552, derived from a soaking experiment | | Descriptor: | (3S,4S,5R)-3-(4-amino-3-{[(2R)-3-ethoxy-1,1,1-trifluoropropan-2-yl]oxy}-5-fluorobenzyl)-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
4D8F
 
 | | Chlamydia trachomatis NrdB with a Mn/Fe cofactor (procedure 1 - high Mn) | | Descriptor: | ACETIC ACID, FE (III) ION, MANGANESE (II) ION, ... | | Authors: | Dassama, L.M.K, Boal, A.K, Krebs, C, Rosenzweig, A.C, Bollinger Jr, J.M. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evidence that the beta subunit of Chlamydia trachomatis ribonucleotide reductase is active with the manganese ion of its manganese(IV)/iron(III) cofactor in site 1.
J.Am.Chem.Soc., 134, 2012
|
|
4Y0A
 
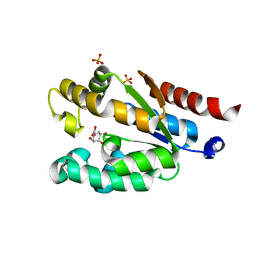 | | Shikimate kinase from Acinetobacter baumannii in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SULFATE ION, Shikimate kinase | | Authors: | Sutton, K.A, Breen, J, MacDonald, U, Beanan, J.M, Olson, R, Russo, T.A, Schultz, L.W, Umland, T.C. | | Deposit date: | 2015-02-05 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Structure of shikimate kinase, an in vivo essential metabolic enzyme in the nosocomial pathogen Acinetobacter baumannii, in complex with shikimate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XUD
 
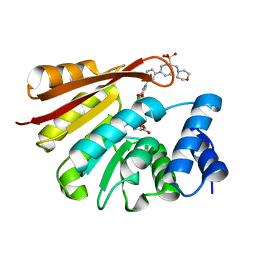 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd32 ([1-(biphenyl-3-yl)-5-hydroxy-4-oxo-1,4-dihydropyridin-3-yl]boronic acid) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4XUL
 
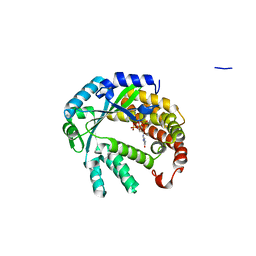 | | Crystal structure of M. chilensis Mg662 protein complexed with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Lartigue, A, Priet, S, Claverie, J.M, Abergel, C. | | Deposit date: | 2015-01-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of M. chilensis Mg662 protein complexed with GTP
To Be Published
|
|
4XKS
 
 | | E. coli BFR variant Y45F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Hemmings, A.M, Le Brun, N.E, Bradley, J.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Three Aromatic Residues are Required for Electron Transfer during Iron Mineralization in Bacterioferritin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XS0
 
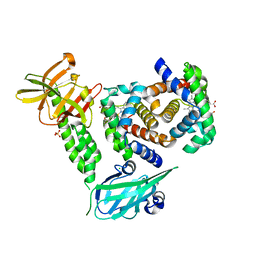 | | Human methemoglobin in complex with the second and third NEAT domains of IsdH(F365Y/A369F/Y642A) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Dickson, C.F, Jacques, D.A, Guss, J.M, Gell, D.A. | | Deposit date: | 2015-01-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of haemoglobin bound to the haemoglobin receptor IsdH from Staphylococcus aureus shows disruption of the native alpha-globin haem pocket.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1JFW
 
 | |
4XWU
 
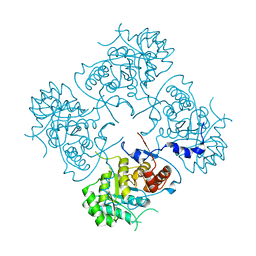 | | Structure of the IMP dehydrogenase from Ashbya gossypii | | Descriptor: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XUE
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd27b | | Descriptor: | 2-(biphenyl-3-yl)-5-hydroxy-3-methylpyrimidin-4(3H)-one, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4D88
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BXQ490 | | Descriptor: | (3S,4S,5R)-3-{4-amino-3-fluoro-5-[(2S)-3,3,3-trifluoro-2-hydroxypropyl]benzyl}-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
