1M35
 
 | | Aminopeptidase P from Escherichia coli | | 分子名称: | AMINOPEPTIDASE P, MANGANESE (II) ION | | 著者 | Graham, S.C, Lee, M, Freeman, H.C, Guss, J.M. | | 登録日 | 2002-06-27 | | 公開日 | 2003-05-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | An orthorhombic form of Escherichia coli aminopeptidase P at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2XU4
 
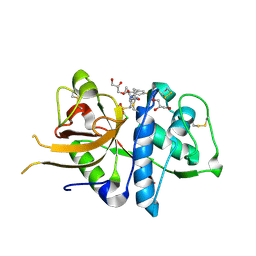 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | 分子名称: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-1-[1-(4-fluorophenyl)cyclopropyl]carbonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | 著者 | Banner, D.W, Benz, J.M, Haap, W. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2XU5
 
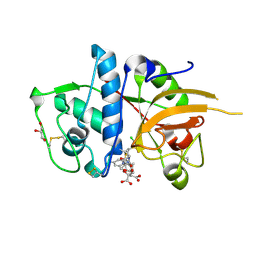 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | 分子名称: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-(4-methylphenyl)cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, CITRATE ANION, ... | | 著者 | Banner, D.W, Benz, J.M, Haap, W. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
1J2O
 
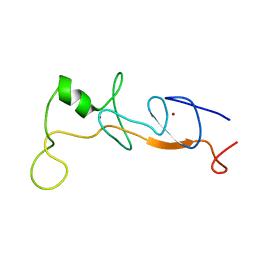 | | Structure of FLIN2, a complex containing the N-terminal LIM domain of LMO2 and ldb1-LID | | 分子名称: | Fusion of Rhombotin-2 and LIM domain-binding protein 1, ZINC ION | | 著者 | Deane, J.E, Mackay, J.P, Kwan, A.H, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | 登録日 | 2003-01-08 | | 公開日 | 2003-05-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
2JTN
 
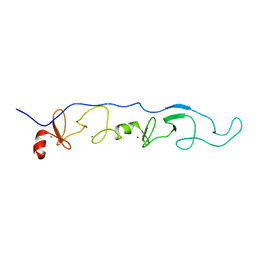 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | 分子名称: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | 著者 | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | 登録日 | 2007-08-03 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
1ML0
 
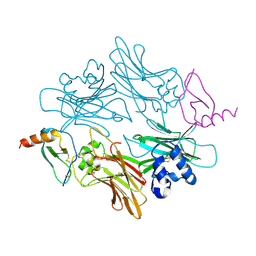 | |
4P0D
 
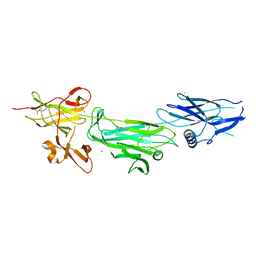 | | The T6 backbone pilin of serotype M6 Streptococcus pyogenes has a modular three-domain structure decorated with variable loops and extensions | | 分子名称: | CALCIUM ION, IODIDE ION, Trypsin-resistant surface T6 protein | | 著者 | Young, P.G, Moreland, N.J, Loh, J.M, Bell, A, Atatoa-Carr, P, Proft, T, Baker, E.N. | | 登録日 | 2014-02-20 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Conservation, Variability, and Immunogenicity of the T6 Backbone Pilin of Serotype M6 Streptococcus pyogenes.
Infect.Immun., 82, 2014
|
|
4OIA
 
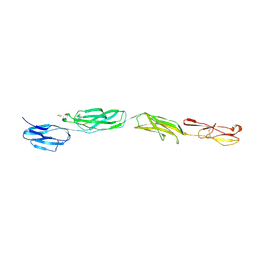 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P4322 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | 登録日 | 2014-01-19 | | 公開日 | 2014-07-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SG3
 
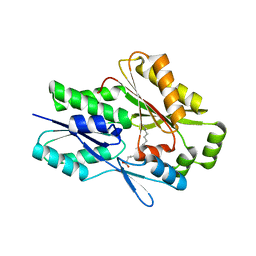 | | The X-ray crystal structure of the Staphylococcus aureus Fatty Acid Kinase B1 mutant A121I-A158L to 2.35 Angstrom resolution (Open form chain A, Palmitate bound) | | 分子名称: | Fatty Acid Kinase B1, PALMITIC ACID | | 著者 | Cuypers, M.G, Gullett, J.M, Rock, C.O, White, S.W. | | 登録日 | 2021-10-04 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7S4Z
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Proteinase K | | 分子名称: | CALCIUM ION, NITRATE ION, Proteinase K | | 著者 | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | 登録日 | 2021-09-09 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S4W
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Lysozyme | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | 登録日 | 2021-09-09 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S4Y
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Insulin | | 分子名称: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | 著者 | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | 登録日 | 2021-09-09 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7SCL
 
 | | The X-ray crystal structure of Staphylococcus aureus Fatty Acid Kinase B1 (FakB1) mutant R173A in complex with Palmitate to 1.60 Angstrom resolution | | 分子名称: | Fatty Acid Kinase B1, GLYCEROL, PALMITIC ACID | | 著者 | Cuypers, M.G, Gullett, J.M, Subramanian, C, Rock, C.O, White, S.W. | | 登録日 | 2021-09-28 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7S56
 
 | | Sortase A from Streptococcus agalactiae, residues 79-247 | | 分子名称: | Class A sortase | | 著者 | Gao, M, Kodama, H.M, Antos, J.M, Amacher, J.F. | | 登録日 | 2021-09-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7S54
 
 | | Sortase A from Streptococcus agalactiae with the deltaN188 b7-b8 loop sequence from Staphylococcus aureus Sortase A | | 分子名称: | Class A sortase, sortase A chimera | | 著者 | Gao, M, Kodama, H.M, Antos, J.M, Amacher, J.F. | | 登録日 | 2021-09-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.794 Å) | | 主引用文献 | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7S57
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence of Enterococcus faecalis Sortase A | | 分子名称: | Class A sortase, sortase A chimera | | 著者 | Svendsen, J.E, Johnson, D.A, Gao, M, Antos, J.M, Amacher, J.F. | | 登録日 | 2021-09-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7S53
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence from Listeria monocytogenes Sortase A | | 分子名称: | Class A sortase, sortase A chimera | | 著者 | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | 登録日 | 2021-09-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | 分子名称: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | 著者 | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | 登録日 | 2021-09-27 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | 分子名称: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | 著者 | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | 登録日 | 2021-09-27 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
4OPX
 
 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2R)-5-fluoro-2-methyl-2,3-dihydro-1-benzofuran-7-carboxamide | | 分子名称: | (2R)-5-fluoro-2-methyl-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | 著者 | Pascal, J.M, Steffen, J.D. | | 登録日 | 2014-02-06 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.314 Å) | | 主引用文献 | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
4OUR
 
 | | Crystal structure of Arabidopsis thaliana phytochrome B photosensory module | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Phytochrome B, ... | | 著者 | Sethe Burgie, E, Bussell, A.N, Walker, J.M, Dubiel, K, Vierstra, R.D. | | 登録日 | 2014-02-18 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OQB
 
 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2Z)-2-{4-[2-(morpholin-4-yl)ethoxy]benzylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide | | 分子名称: | (2Z)-2-{4-[2-(morpholin-4-yl)ethoxy]benzylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | 著者 | Pascal, J.M, Steffen, J.D. | | 登録日 | 2014-02-07 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.362 Å) | | 主引用文献 | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
7SJJ
 
 | | Crystal structure of photoactive yellow protein (PYP); F96oCNF construct | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Weaver, J.B, Kirsh, J.M, Boxer, S.G. | | 登録日 | 2021-10-17 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments.
J.Am.Chem.Soc., 144, 2022
|
|
7S81
 
 | | Structure of human PARP1 domains (Zn1, Zn3, WGR, HD) bound to a DNA double strand break. | | 分子名称: | DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*CP*GP*CP*AP*T)-3'), Poly [ADP-ribose] polymerase 1, ZINC ION | | 著者 | Rouleau-Turcotte, E, Pascal, J.M. | | 登録日 | 2021-09-17 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S6H
 
 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | 分子名称: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | 著者 | Rouleau-Turcotte, E, Pascal, J.M. | | 登録日 | 2021-09-14 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
