8V5R
 
 | | Active conformation of DNA polymerase gamma bound to DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, ... | | Authors: | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, J.M, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
4WTW
 
 | | Crystal structure of the third FnIII domain of integrin beta4 | | Descriptor: | 1,2-ETHANEDIOL, Integrin beta-4, PENTAETHYLENE GLYCOL, ... | | Authors: | Alonso-Garcia, N, Urien, H, de Pereda, J.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin alpha6beta4
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6PCY
 
 | |
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
4WST
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSW
 
 | | The crystal structure of hemagglutinin from A/green-winged teal/Texas/Y171/2006 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
1ERI
 
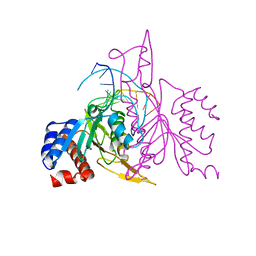 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
4XBU
 
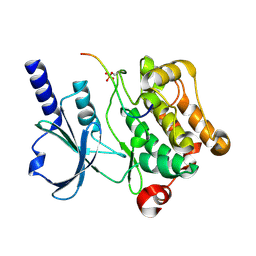 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | Descriptor: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XGC
 
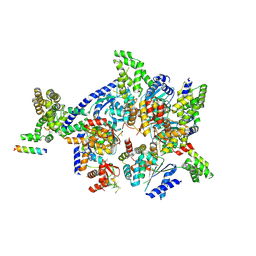 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
1EBK
 
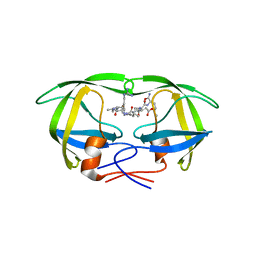 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
6PTL
 
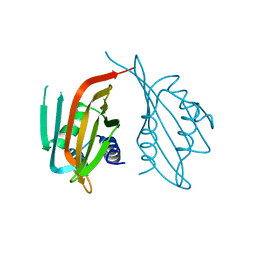 | |
6ZOW
 
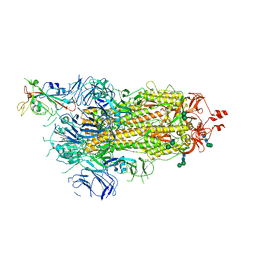 | | SARS-CoV-2 spike in prefusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZP5
 
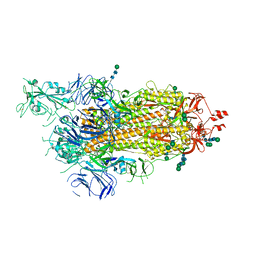 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
1ELS
 
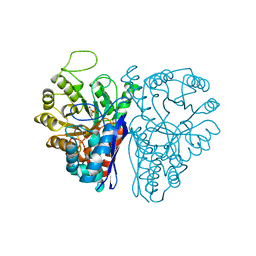 | | CATALYTIC METAL ION BINDING IN ENOLASE: THE CRYSTAL STRUCTURE OF ENOLASE-MN2+-PHOSPHONOACETOHYDROXAMATE COMPLEX AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, MANGANESE (II) ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Zhang, E, Hatada, M, Brewer, J.M, Lebioda, L. | | Deposit date: | 1994-04-05 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic metal ion binding in enolase: the crystal structure of an enolase-Mn2+-phosphonoacetohydroxamate complex at 2.4-A resolution.
Biochemistry, 33, 1994
|
|
5NJO
 
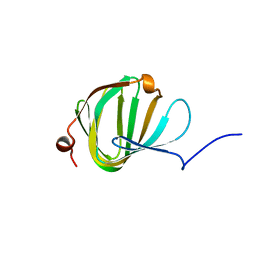 | | Roll out the beta-barrel: structure and mechanism of Pac13, a unique nucleoside dehydratase | | Descriptor: | Putative cupin_2 domain-containing isomerase | | Authors: | Michailidou, F, Bent, A.F, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2017-03-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1EV1
 
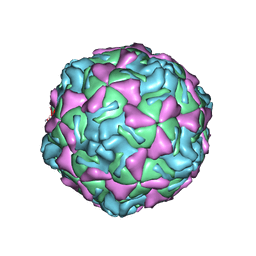 | | ECHOVIRUS 1 | | Descriptor: | ECHOVIRUS 1, MYRISTIC ACID, PALMITIC ACID | | Authors: | Wien, M.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-12-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure determination of echovirus 1.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6R95
 
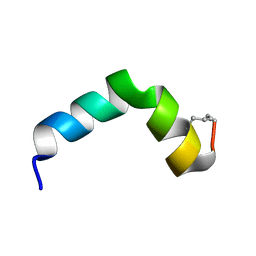 | |
6R96
 
 | |
7YR6
 
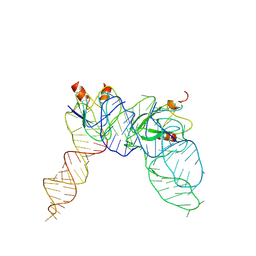 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers | | Descriptor: | RsmZ RNA, Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Ling, X, Yang, X, Wu, Y, Liu, T, Wei, X, Bujnick, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
8SLZ
 
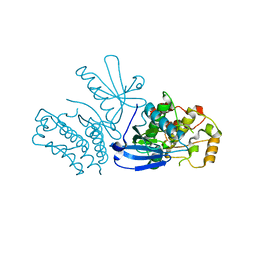 | |
5Y22
 
 | | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV | | Descriptor: | 22AA-PSTD peptide | | Authors: | Lu, B, Liao, S.M, Huang, J.M, Lu, Z.L, Chen, D, Liu, X.H, Zhou, G.P, Huang, R.B. | | Deposit date: | 2017-07-23 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV
To Be Published
|
|
5DS3
 
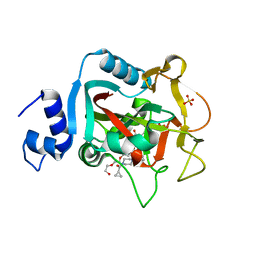 | | Crystal structure of constitutively active PARP-1 | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, PENTAETHYLENE GLYCOL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
5DSY
 
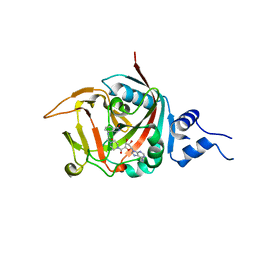 | | Crystal structure of constitutively active PARP-2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 2 | | Authors: | Riccio, A.A, Pascal, J.M. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
5ZRT
 
 | | Crystal structure of human C1ORF123 protein | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rahaman, S.N.A, Yusop, J.M, Mohamed-Hussein, Z.A, Wan Mohd, A, Ho, K.L, Teh, A.H, Waterman, J, Ng, C.L. | | Deposit date: | 2018-04-25 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of human C1ORF123.
Peerj, 6, 2018
|
|
5FQM
 
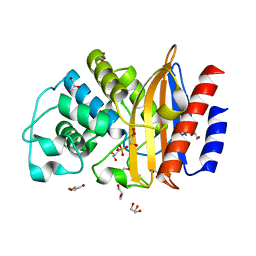 | | Last common ancestor of Gram Negative Bacteria (GNCA) Class A beta- lactamase | | Descriptor: | GLYCEROL, GNCA BETA LACTAMASE, SULFATE ION | | Authors: | Martinez Rodriguez, S, Gavira, J.A, Risso, V.A, Sanchez Ruiz, J.M. | | Deposit date: | 2015-12-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
