1G10
 
 | | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR PROTEIN NMR STRUCTURE | | Descriptor: | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR | | Authors: | Hemmi, H, Studts, J.M, Chae, Y.K, Song, J, Markley, J.L, Fox, B.G. | | Deposit date: | 2000-10-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the toluene 4-monooxygenase effector protein (T4moD).
Biochemistry, 40, 2001
|
|
6OEF
 
 | | PolyAla Model of the O-layer from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of OMCC O-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
8ECO
 
 | | Microbacterium phage Oxtober96 | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EC2
 
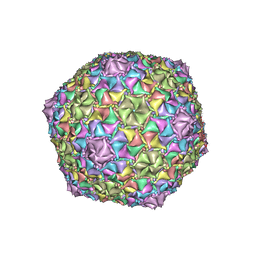 | | Mycobacterium phage Adephagia | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
1CDB
 
 | |
8ECJ
 
 | | Mycobacterium phage Cain | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECN
 
 | | Mycobacterium phage Ogopogo | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EB4
 
 | | Gordonia phage Ziko | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-08-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECI
 
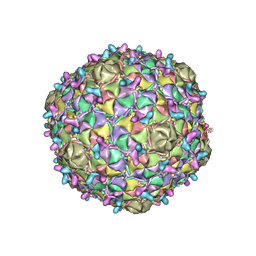 | | Arthrobacter phage Bridgette | | Descriptor: | Decoration protein, Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EC8
 
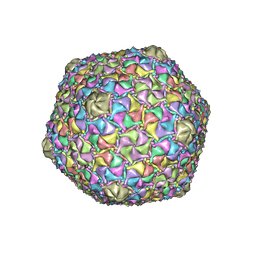 | | Mycobacterium phage Bobi | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8E16
 
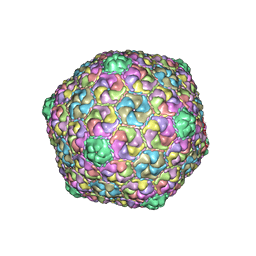 | | Mycobacterium phage Che8 | | Descriptor: | Major capsid protein, gp6 | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECK
 
 | | Gordonia phage Cozz | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
6OW4
 
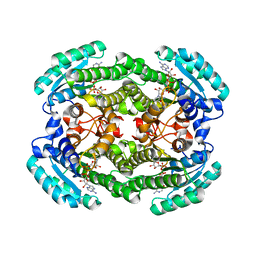 | | Structure of the NADH-bound form of 20beta-Hydroxysteroid Dehydrogenase from Bifidobacterium adolescentis strain L2-32 | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Mythen, S.M, Pollet, R.M, Koropatkin, N.M, Ridlon, J.M. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and biochemical characterization of 20 beta-hydroxysteroid dehydrogenase fromBifidobacterium adolescentisstrain L2-32.
J.Biol.Chem., 294, 2019
|
|
1BQ0
 
 | |
6AY5
 
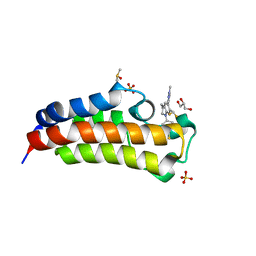 | | CREBBP bromodomain in complex with Cpd17 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-3-methylbenzo[d]thiazol-2(3H)-one) | | Descriptor: | 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-3-methyl-1,3-benzothiazol-2(3H)-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
4YPI
 
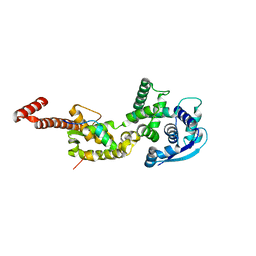 | | Structure of Ebola virus nucleoprotein N-terminal fragment bound to a peptide derived from Ebola VP35 | | Descriptor: | Nucleoprotein, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Borek, D.M, Binning, J.M, Otwinowski, Z, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.
Cell Rep, 11, 2015
|
|
4YUK
 
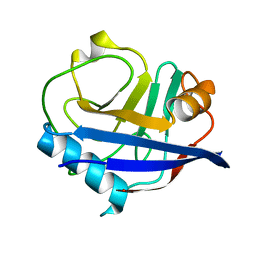 | | Multiconformer synchrotron model of CypA at 260 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
8E17
 
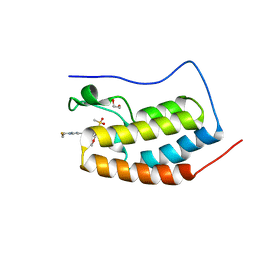 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6M)-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-4-[5-(methanesulfonyl)-2-methoxyphenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-09 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8E3W
 
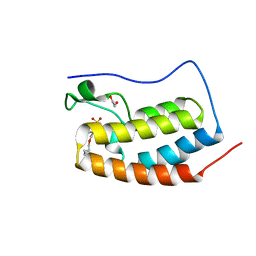 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8DYR
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
4YGL
 
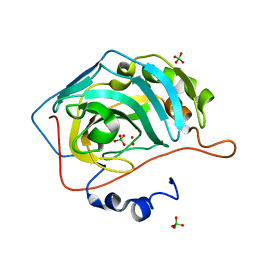 | | NaClO4--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | Carbonic anhydrase 2, HYDROXIDE ION, PERCHLORATE ION, ... | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
7R4Q
 
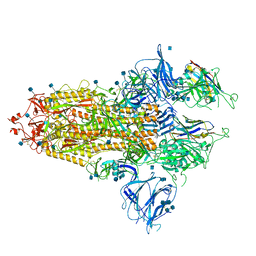 | | The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.29, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4I
 
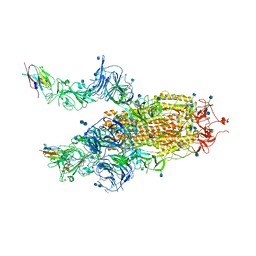 | | The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 2.15, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4R
 
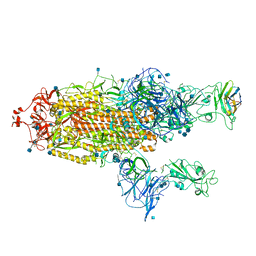 | | The SARS-CoV-2 spike in complex with the 1.10 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.10, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7CUE
 
 | | Crystal structure of HID2 bound to human Hemoglobin | | Descriptor: | Amino acid ABC transporter substrate-binding protein, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Caaveiro, J.M.M, Hoshino, M, Tsumoto, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the recognition of human hemoglobin by the Shr protein from Streptococcus pyogenes
To Be Published
|
|
