8BX8
 
 | | In situ outer dynein arm from Chlamydomonas reinhardtii in the post-power stroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Zimmermann, N.E.L, Noga, A, Obbineni, J.M, Ishikawa, T. | | Deposit date: | 2022-12-08 | | Release date: | 2023-05-10 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (30.299999 Å) | | Cite: | ATP-induced conformational change of axonemal outer dynein arms revealed by cryo-electron tomography.
Embo J., 42, 2023
|
|
1UDU
 
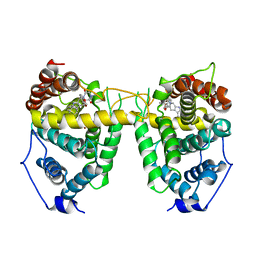 | | Crystal structure of Human Phosphodiesterase 5 complexed with tadalafil(Cialis) | | Descriptor: | 6-BENZO[1,3]DIOXOL-5-YL-2-METHYL-2,3,6,7,12,12A-HEXAHYDRO-PYRAZINO[1',2':1,6]PYRIDO[3,4-B]INDOLE-1,4-DIONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Sung, B.-J, Lee, J.I, Heo, Y.-S, Kim, J.H, Moon, J, Yoon, J.M, Hyun, Y.-L, Kim, E, Eum, S.J, Lee, T.G, Cho, J.M, Park, S.-Y, Lee, J.-O, Jeon, Y.H, Hwang, K.Y, Ro, S. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules
Nature, 425, 2003
|
|
8JJG
 
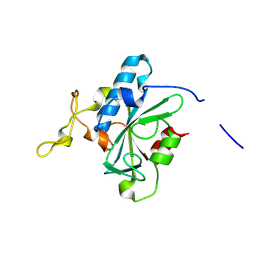 | | Crystal structure of QW-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJI
 
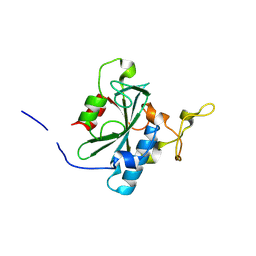 | | Crystal structure of QR-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK1
 
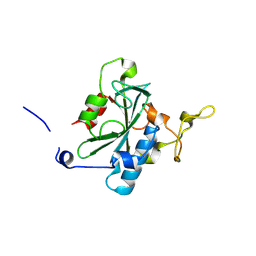 | | Crystal structure of QA-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJH
 
 | | Crystal structure of QH-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK0
 
 | | Crystal structure of QL-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJU
 
 | | Crystal structure of QD-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJY
 
 | | Crystal structure of QN-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJW
 
 | | Crystal structure of QG-hNTAQ1 C28S | | Descriptor: | MAGNESIUM ION, Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJZ
 
 | | Crystal structure of QQ-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJX
 
 | | Crystal structure of QS-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJF
 
 | | Crystal structure of QE-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK2
 
 | | Crystal structure of QF-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
6D43
 
 | |
8P1J
 
 | |
8P1L
 
 | |
8OJS
 
 | | Crystal structure of the human IgD Fab - structure Fab1 | | Descriptor: | 1,2-ETHANEDIOL, Human IgD Fab heavy chain, Human IgD Fab light chain, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJU
 
 | | Crystal structure of the human IgD Fab - structure Fab3 | | Descriptor: | 1,2-ETHANEDIOL, Human IgD Fab heavy chain, Human IgD Fab light chain, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJV
 
 | | Crystal structure of the human IgD Fab - structure Fab4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJT
 
 | | Crystal structure of the human IgD Fab - structure Fab2 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OXL
 
 | |
8P3W
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 4 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3X
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8OXK
 
 | |
