5QD6
 
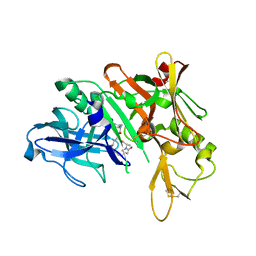 | | Crystal structure of BACE complex with BMC004 | | Descriptor: | (3S,14R,16S)-16-[1,1-dihydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5GWF
 
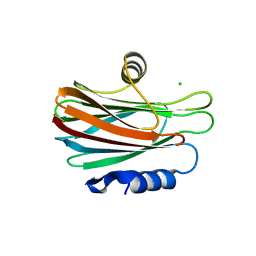 | | FraC with GlcNAc(6S) bound | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, CHLORIDE ION, DELTA-actitoxin-Afr1a, ... | | Authors: | Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2016-09-11 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Haemolytic actinoporins interact with carbohydrates using their lipid-binding module
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
5YQ5
 
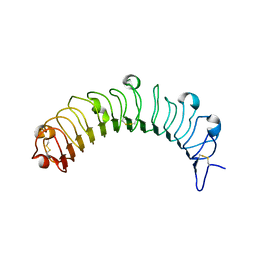 | | Crystal structure of human osteomodulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Osteomodulin, PHOSPHATE ION | | Authors: | Caaveiro, J.M.M, Tashima, T, Tsumoto, K. | | Deposit date: | 2017-11-05 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular basis for governing the morphology of type-I collagen fibrils by Osteomodulin.
Commun Biol, 1, 2018
|
|
1U68
 
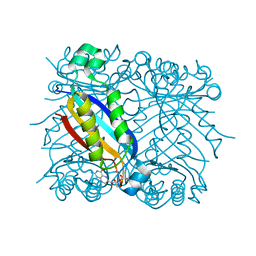 | | DHNA 7,8 DIHYDRONEOPTERIN COMPLEX | | Descriptor: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent inhibitors of dihydroneopterin aldolase using CrystaLEAD high-throughput X-ray crystallographic screening and structure-directed lead optimization.
J.MED.CHEM., 47, 2004
|
|
5QD1
 
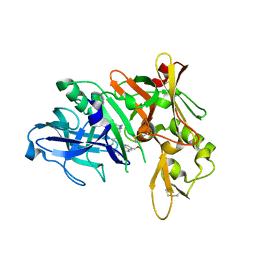 | | Crystal structure of BACE complex with BMC011 | | Descriptor: | (10S,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-7-oxa-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1UB1
 
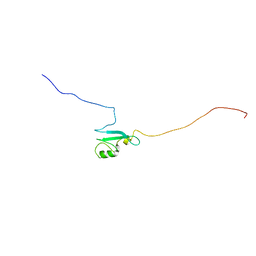 | | Solution structure of the matrix attachment region-binding domain of chicken MeCP2 | | Descriptor: | attachment region binding protein | | Authors: | Heitmann, B, Maurer, T, Weitzel, J.M, Stratling, W.H, Kalbitzer, H.R, Brunner, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the matrix attachment region-binding domain of chicken MeCP2
EUR.J.BIOCHEM., 270, 2003
|
|
5QCZ
 
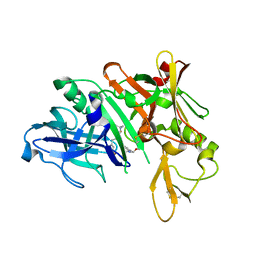 | | Crystal structure of BACE complex with BMC015 | | Descriptor: | (4S)-4-{(S)-hydroxy[(3R,6R)-6-(methoxymethyl)morpholin-3-yl]methyl}-19-(methoxymethyl)-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1TVK
 
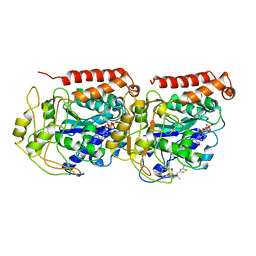 | | The binding mode of epothilone A on a,b-tubulin by electron crystallography | | Descriptor: | EPOTHILONE A, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nettles, J.H, Li, H, Cornett, B, Krahn, J.M, Snyder, J.P, Downing, K.H. | | Deposit date: | 2004-06-29 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.89 Å) | | Cite: | The binding mode of epothilone A on alpha,beta-tubulin by electron crystallography
Science, 305, 2004
|
|
5QCT
 
 | | Crystal structure of BACE complex with BMC001 | | Descriptor: | (2R,4S)-N-butyl-4-[(4S,6R)-16-ethoxy-12-ethyl-6-methyl-2,13-dioxo-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-trien-4-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1, PHOSPHATE ION | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1UCI
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
5YD3
 
 | | Crystal structure of the scFv antibody 4B08 with epitope peptide | | Descriptor: | Epitope peptide, GLYCEROL, SULFATE ION, ... | | Authors: | Caaveiro, J.M.M, Miyanabe, K, Tsumoto, K. | | Deposit date: | 2017-09-11 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Intramolecular H-bonds govern the recognition of a flexible peptide by an antibody
J. Biochem., 164, 2018
|
|
5QCR
 
 | | Crystal structure of BACE complex with BMC026 | | Descriptor: | 2-(butylamino)-N-[(2S,3S,5R)-6-(butylamino)-3-hydroxy-5-methyl-6-oxo-1-phenylhexan-2-yl]-6-methoxypyridine-4-carboxamide, Beta-secretase 1, SULFATE ION | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5YC5
 
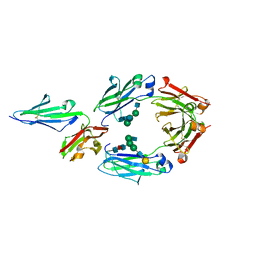 | | Crystal structure of human IgG-Fc in complex with aglycan and optimized Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Tsumoto, K, Kiyoshi, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Assessing the Heterogeneity of the Fc-Glycan of a Therapeutic Antibody Using an engineered Fc gamma Receptor IIIa-Immobilized Column.
Sci Rep, 8, 2018
|
|
5QD0
 
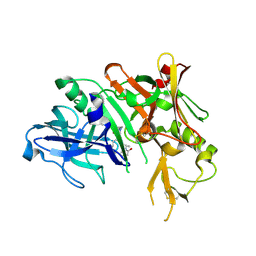 | | Crystal structure of BACE complex withBMC006 | | Descriptor: | (5S,8S,10R)-8-[(1R)-1-hydroxy-2-{[(5-propyl-1H-pyrazol-3-yl)methyl]amino}ethyl]-4,5,10-trimethyl-1-oxa-4,7-diazacyclohexadecane-3,6-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
8FHU
 
 | |
8FHL
 
 | |
5QCV
 
 | | Crystal structure of BACE complex with BMC023 | | Descriptor: | (10S,13S)-13-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9,10-dimethyl-2-oxa-9,12-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-8,11-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6AFB
 
 | | DJ-1 C106S incubated with isatin | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, ISATIN, ... | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFL
 
 | | DJ-1 with compound 15 | | Descriptor: | 5-fluoranyl-1-(2-phenylethyl)indole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFD
 
 | | DJ-1 with compound 6 | | Descriptor: | 7-methyl-1~{H}-indole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFC
 
 | | DJ-1 with compound 4 | | Descriptor: | 5-fluoranyl-1~{H}-indole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AF7
 
 | | DJ-1 C106S unbound | | Descriptor: | CHLORIDE ION, PENTAETHYLENE GLYCOL, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFI
 
 | | DJ-1 with compound 11 | | Descriptor: | 1-ethylindole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFH
 
 | | DJ-1 with compound 10 | | Descriptor: | 1-(2-phenylethyl)indole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AF5
 
 | | DJ-1 after backsoaking | | Descriptor: | CHLORIDE ION, ISATIN, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
