2JE7
 
 | | Crystal structure of recombinant Dioclea guianensis lectin S131H complexed with 5-bromo-4-chloro-3-indolyl-a-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, LECTIN ALPHA CHAIN, ... | | Authors: | Nagano, C.S, Sanz, L, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights Into the Structural Basis of the Ph- Dependent Dimer-Tetramer Equilibrium Through Crystallographic Analysis of Recombinant Diocleinae Lectins.
Biochem.J., 409, 2008
|
|
2ORM
 
 | | Crystal Structure of the 4-Oxalocrotonate Tautomerase Homologue DmpI from Helicobacter pylori. | | Descriptor: | Probable tautomerase HP0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Czerwinski, R.M, Kern, A.D. | | Deposit date: | 2007-02-03 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
6JPQ
 
 | | CryoEM structure of Abo1 hexamer - ADP complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
6JFK
 
 | | GDP bound Mitofusin2 (MFN2) | | Descriptor: | CITRIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
6KP6
 
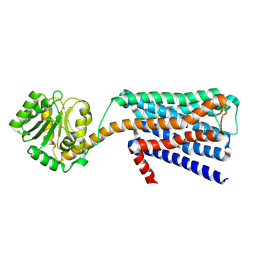 | | The structural study of mutation induced inactivation of Human muscarinic receptor M4 | | Descriptor: | Muscarinic acetylcholine receptor M4,GlgA glycogen synthase,Muscarinic acetylcholine receptor M4 | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural study of mutation-induced inactivation of human muscarinic receptor M4
Iucrj, 7, 2020
|
|
2JVU
 
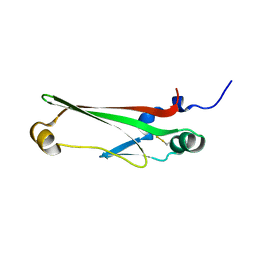 | | Solution Structure of Dispersin from Enteroaggregative Escherichia coli | | Descriptor: | DISPERSIN | | Authors: | Velarde, J.J, Varney, K.M, Farfan, K, Dudley, D, Inman, J.G, Fletcher, J, Weber, D.J, Nataro, J.P. | | Deposit date: | 2007-09-25 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the novel dispersin protein of enteroaggregative Escherichia coli.
Mol.Microbiol., 66, 2007
|
|
2LJC
 
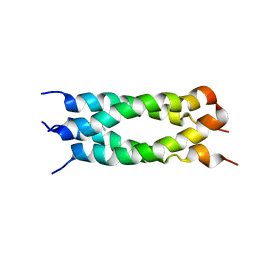 | |
6KVN
 
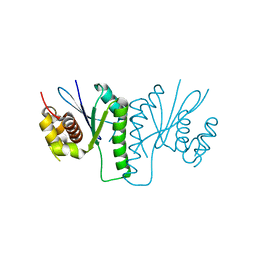 | |
2JQ9
 
 | | VPS4A MIT-CHMP1A complex | | Descriptor: | Chromatin-modifying protein 1a, Vacuolar protein sorting-associating protein 4A | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-05-30 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2ONX
 
 | | NNQQ peptide corresponding to residues 8-11 of yeast prion sup35 (alternate crystal form) | | Descriptor: | peptide corresponding to residues 8-11 of yeast prion sup35 | | Authors: | Sawaya, M.R, Sambashivan, S, Nelson, R, Ivanova, M, Sievers, S.A, Apostol, M.I, Thompson, M.J, Balbirnie, M, Wiltzius, J.J, McFarlane, H, Madsen, A.O, Riekel, C, Eisenberg, D. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Atomic structures of amyloid cross-beta spines reveal varied steric zippers.
Nature, 447, 2007
|
|
2OP8
 
 | |
2OLQ
 
 | | How Does an Enzyme Recognize CO2? | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBON DIOXIDE, MAGNESIUM ION, ... | | Authors: | Puttick, J, Goldie, H, Cotelesage, J.J, Rajabi, B, Novakovski, B, Delbaere, L.T. | | Deposit date: | 2007-01-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | How does an enzyme recognize CO2?
Int.J.Biochem.Cell Biol., 39, 2007
|
|
6I90
 
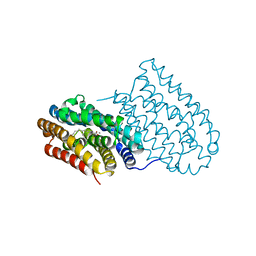 | |
2KTZ
 
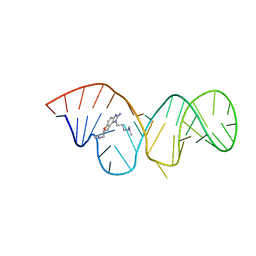 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7R)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2GQQ
 
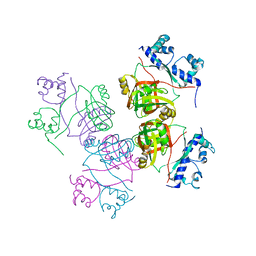 | |
2K6T
 
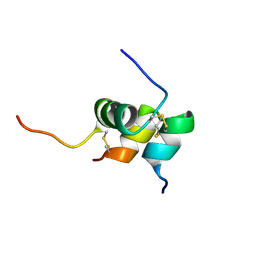 | | Solution structure of the relaxin-like factor | | Descriptor: | Insulin-like 3 A chain, Insulin-like 3 B chain | | Authors: | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
2JE9
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT DIOCLEA GRANDIFLORA LECTIN COMPLEXED WITH 5-BROMO-4-CHLORO-3-INDOLYL-A-D-MANNOSE | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, LECTIN ALPHA CHAIN, ... | | Authors: | Nagano, C.S, Sanz, L, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2007-01-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structural Basis of the Ph- Dependent Dimer-Tetramer Equilibrium Through Crystallographic Analysis of Recombinant Diocleinae Lectins.
Biochem.J., 409, 2008
|
|
2JEC
 
 | | crystal structure of recombinant DiocleA grandiflora lectin mutant E123A-H131N-K132Q complexed witH 5-bromo-4-chloro-3-indolyl-a-D- mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, LECTIN ALPHA CHAIN, ... | | Authors: | Nagano, C.S, Sanz, L, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2007-01-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights Into the Structural Basis of the Ph- Dependent Dimer-Tetramer Equilibrium Through Crystallographic Analysis of Recombinant Diocleinae Lectins.
Biochem.J., 409, 2008
|
|
2HHW
 
 | | ddTTP:O6-methyl-guanine pair in the polymerase active site, in the closed conformation | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HJK
 
 | | Crystal Structure of HLA-B5703 and HIV-1 peptide | | Descriptor: | Beta-2-microglobulin, Gag protein, HLA class I histocompatibility antigen B-57 | | Authors: | Gillespie, G.M.A, Stewart-Jones, G, Rengasamy, J, Beattie, T, Bwayo, J.J, Plummer, F.A. | | Deposit date: | 2006-06-30 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Strong TCR Conservation and Altered T Cell Cross-Reactivity Characterize a B*57-Restricted Immune Response in HIV-1 Infection.
J.Immunol., 177, 2006
|
|
2HHS
 
 | | O6-methyl:C pair in the polymerase-10 basepair position | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*GP*CP*AP*TP*GP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HT8
 
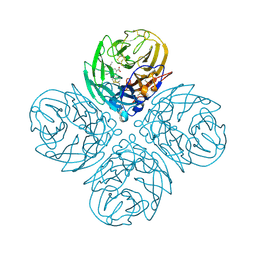 | | N8 neuraminidase in complex with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2HLG
 
 | |
6LCT
 
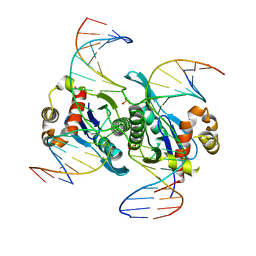 | | Crystal structure of catalytic inactive chloroplast resolvase NtMOC1 in complex with Holliday junction | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*GP*TP*CP*CP*CP*TP*CP*TP*GP*TP*TP*GP*T)-3'), DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*GP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*GP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), ... | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
2HTV
 
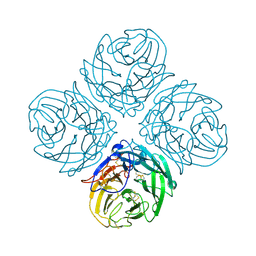 | | N4 neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
