2NA6
 
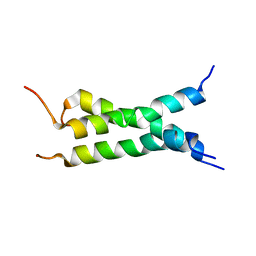 | | Transmembrane domain of mouse Fas/CD95 death receptor | | Descriptor: | Tumor necrosis factor receptor superfamily member 6 | | Authors: | Fu, Q, Chou, J.J, Wu, H, Fu, T, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis and Functional Role of Intramembrane Trimerization of the Fas/CD95 Death Receptor.
Mol.Cell, 61, 2016
|
|
2FZM
 
 | |
2G8S
 
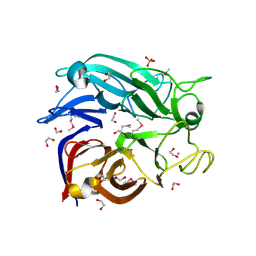 | | Crystal structure of the soluble Aldose sugar dehydrogenase (Asd) from Escherichia coli in the apo-form | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/sorbosone dehydrogenases, ... | | Authors: | Southall, S.M, Doel, J.J, Richardson, D.J, Oubrie, A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Soluble Aldose Sugar Dehydrogenase from Escherichia coli: A HIGHLY EXPOSED ACTIVE SITE CONFERRING BROAD SUBSTRATE SPECIFICITY.
J.Biol.Chem., 281, 2006
|
|
2NRZ
 
 | |
2J6R
 
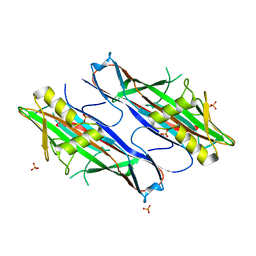 | | FaeG from F4ac ETEC strain GIS26, produced in tobacco plant chloroplast | | Descriptor: | K88 FIMBRIAL PROTEIN, PHOSPHATE ION | | Authors: | Van Molle, I, Joensuu, J.J, Buts, L, Panjikar, S, Kotiaho, M, Bouckaert, J, Wyns, L, Niklander-Teeri, V, De Greve, H. | | Deposit date: | 2006-10-03 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chloroplasts Assemble the Major Subunit Faeg of Escherichia Coli F4 (K88) Fimbriae Into Strand-Swapped Dimers
J.Mol.Biol., 368, 2007
|
|
5KF7
 
 | |
8OUR
 
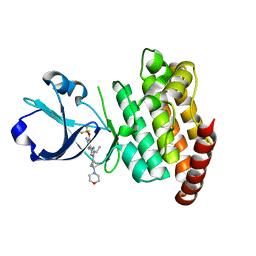 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 5-[1-(3-morpholin-4-yl-1-bicyclo[1.1.1]pentanyl)-2-propan-2-yl-imidazol-4-yl]-3-(trifluoromethyloxy)pyridin-2-amine, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, McEwan, P.A, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
8OUT
 
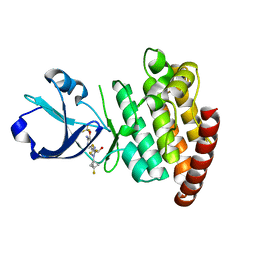 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 22 | | Descriptor: | (1R)-1-[4-[6-azanyl-5-(trifluoromethyloxy)pyridin-3-yl]-1-(3-fluoranyl-1-bicyclo[1.1.1]pentanyl)imidazol-2-yl]-2,2,2-tris(fluoranyl)ethanol, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, Akkermans, O, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
8OW4
 
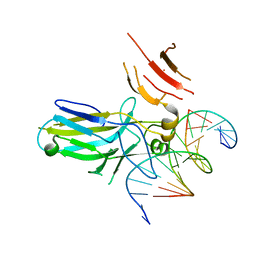 | | 2.75 angstrom crystal structure of human NFAT1 with bound DNA | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*TP*TP*CP*CP*AP*GP*C)-3'), DNA (5'-D(*TP*TP*GP*CP*TP*GP*GP*AP*AP*AP*AP*AP*TP*AP*G)-3'), Nuclear factor of activated T-cells, ... | | Authors: | Lopez-Sagaseta, J, Erausquin, E, Hernandez-Morales, S, Urdiciain, A, Lasarte, J.J, Lozano, T. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 angstrom crystal structure of human NFAT1 with bound DNA
Not published
|
|
5JCD
 
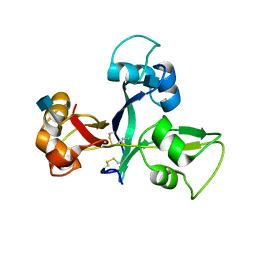 | | Crystal structure of OsCEBiP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor-binding protein | | Authors: | Chai, J.J, Liu, S.M, Wang, J.Z. | | Deposit date: | 2016-04-15 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP
Structure, 24, 2016
|
|
8OUS
 
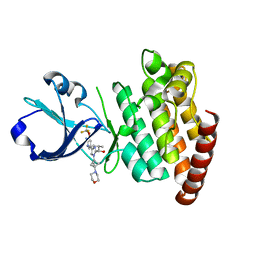 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 19 | | Descriptor: | (1S)-1-[4-[6-azanyl-5-(trifluoromethyloxy)pyridin-3-yl]-1-(3-morpholin-4-yl-1-bicyclo[1.1.1]pentanyl)imidazol-2-yl]-2-methyl-propan-1-ol, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, McEwan, P.A, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
2O1W
 
 | |
6I92
 
 | |
2G7O
 
 | | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM | | Descriptor: | Protein traM | | Authors: | Lu, J, Edwards, R.A, Wong, J.J, Manchak, J, Scott, P.G, Frost, L.S, Glover, J.N. | | Deposit date: | 2006-02-28 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM.
Embo J., 25, 2006
|
|
2NA7
 
 | |
2NQO
 
 | | Crystal Structure of Helicobacter pylori gamma-Glutamyltranspeptidase | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Boanca, G, Sand, A, Okada, T, Suzuki, H, Kumagai, H, Fukuyama, K, Barycki, J.J. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad.
J.Biol.Chem., 282, 2007
|
|
2FZN
 
 | |
2N4X
 
 | |
2O8D
 
 | | human MutSalpha (MSH2/MSH6) bound to ADP and a G dU mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*(DU)P*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2OBE
 
 | | Crystal Structure of Chimpanzee Adenovirus (Type 68/Simian 25) Major Coat Protein Hexon | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIHYDROGENPHOSPHATE ION, Hexon protein | | Authors: | Xue, F, Rux, J.J, Burnett, R.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based identification of a major neutralizing site in an adenovirus hexon
J.Virol., 81, 2007
|
|
2NR1
 
 | | TRANSMEMBRANE SEGMENT 2 OF NMDA RECEPTOR NR1, NMR, 10 STRUCTURES | | Descriptor: | NR1 M2 | | Authors: | Gesell, J.J, Sun, W, Montal, M, Opella, S. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
2O4E
 
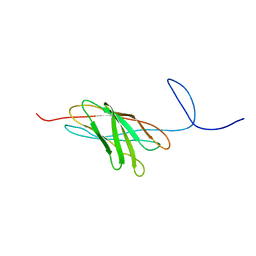 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
2OLR
 
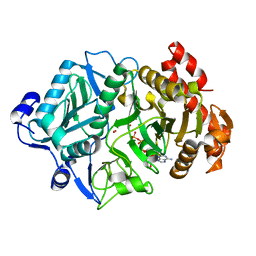 | | Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase complexed with carbon dioxide, Mg2+, ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Cotelesage, J.J, Delbaere, L.T, Goldie, H, Puttick, J, Rajabi, B, Novakovski, B. | | Deposit date: | 2007-01-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How does an enzyme recognize CO2?
Int.J.Biochem.Cell Biol., 39, 2007
|
|
2JDZ
 
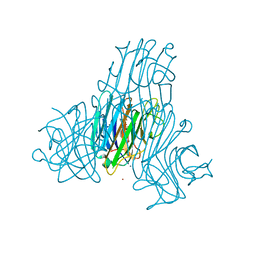 | | Crystal structure of recombinant Dioclea guianensis lectin complexed with 5-bromo-4-chloro-3-indolyl-a-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CADMIUM ION, CALCIUM ION, ... | | Authors: | Nagano, C.S, Sanz, L, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2007-01-12 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structural Basis of the Ph-Dependent Dimer-Tetramer Equilibrium Through Crystallographic Analysis of Recombinant Diocleinae Lectins.
Biochem.J., 409, 2008
|
|
2OPA
 
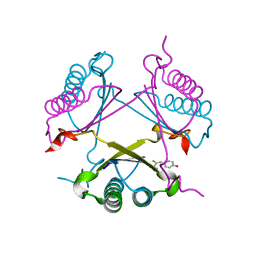 | | YwhB binary complex with 2-Fluoro-p-hydroxycinnamate | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, Probable tautomerase ywhB | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J. | | Deposit date: | 2007-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of YwhB, a 4-Oxalocrotonate Tautomerase Homologue from Bacillus subtilis: the Structural Basis for Catalysis, Inhibition, and Reaction Stereoselectivity.
TO BE PUBLISHED
|
|
