4Q3X
 
 | |
7RVG
 
 | |
6R7O
 
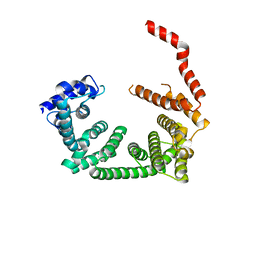 | | Crystal structure of the central region of human cohesin subunit STAG1 | | Descriptor: | Cohesin subunit SA-1 | | Authors: | Newman, J.A, katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-03-29 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
7RWP
 
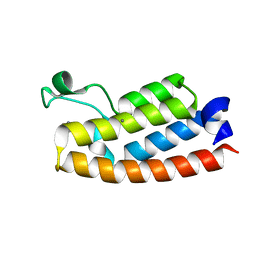 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | Descriptor: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
1CVA
 
 | |
7RVI
 
 | |
4Q37
 
 | |
7RVL
 
 | |
7RVC
 
 | |
7RVE
 
 | |
7RVF
 
 | |
4Q9R
 
 | | Crystal structure of an RNA aptamer bound to trifluoroethyl-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
7RVJ
 
 | |
7RVK
 
 | |
7RWQ
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
6R5R
 
 | |
8BD0
 
 | |
4QOJ
 
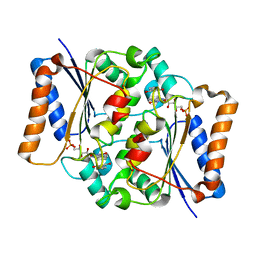 | | CRYSTAL STRUCTURE OF FMN QUINONE REDUCTASE 2 IN COMPLEX WITH RESVERATROL AT 1.85A | | Descriptor: | FLAVIN MONONUCLEOTIDE, RESVERATROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | CRYSTAL STRUCTURE OF FMN QUINONE REDUCTASE 2 IN COMPLEX WITH RESVERATROL AT 1.85A
To be Published
|
|
6RBJ
 
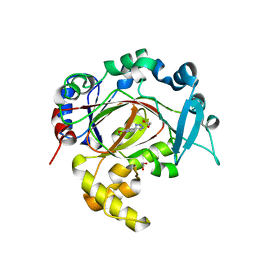 | | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-1,2,3,4-tetrazol-5-yl)quinolin-8-ol, CHLORIDE ION, ... | | Authors: | Johansson, C, Newman, J.A, Kawamura, A, Schofield, C.J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol
To Be Published
|
|
1CVC
 
 | |
8BPI
 
 | |
8BLX
 
 | | Engineered Fructosyl Peptide Oxidase - X02A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (X02A), ... | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
8BJY
 
 | | Engineered Fructosyl Peptide Oxidase - X02B mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (X02B), GLYCEROL | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
8BLZ
 
 | | Engineered Fructosyl Peptide Oxidase - D02 mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (D02), GLYCEROL, ... | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
