4AEK
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4BZJ
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4BPY
 
 | |
4AEM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
4ARD
 
 | | Structure of the immature retroviral capsid at 8A resolution by cryo- electron microscopy | | Descriptor: | CAPSID PROTEIN P27 | | Authors: | Bharat, T.A.M, Davey, N.E, Ulbrich, P, Riches, J.D, Marco, A.D, Rumlova, M, Sachse, C, Ruml, T, Briggs, J.A.G. | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structure of the Immature Retroviral Capsid at 8A Resolution by Cryo-Electron Microscopy.
Nature, 487, 2012
|
|
4BBO
 
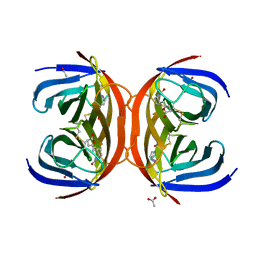 | | Crystal structure of core-bradavidin | | Descriptor: | ACETATE ION, BIOTIN, BLR5658 PROTEIN, ... | | Authors: | Airenne, T.T, Johnson, M.S, Maatta, J.A.E, Hytonen, V.H, Kulomaa, M.S. | | Deposit date: | 2012-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Core-Bradavidin
To be Published
|
|
4AFD
 
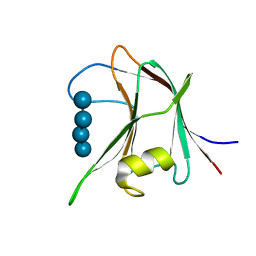 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens with a partially bound cellotetraose moeity. | | Descriptor: | ENDOGLUCANASE CEL5A, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Understanding How Noncatalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4CDX
 
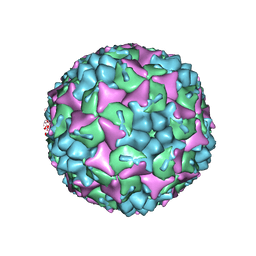 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | Descriptor: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDW
 
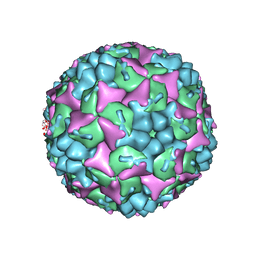 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | Descriptor: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
3ZNU
 
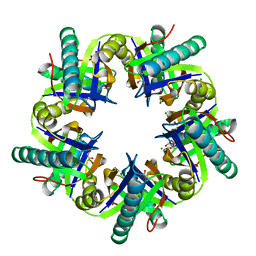 | | Crystal structure of ClcF in crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION, ... | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
3ZOO
 
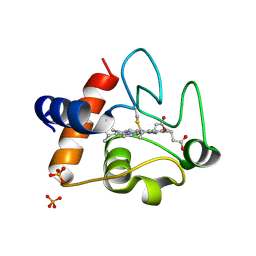 | |
3ZCF
 
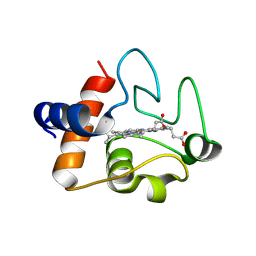 | | Structure of recombinant human cytochrome c | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Rajagopal, B.S, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2012-11-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Hydrogen Peroxide Induced Radical Behaviour in Human Cytochrome C Phospholipid Complexes: Implications for the Enhanced Pro-Apoptotic Activity of the G41S Mutant
Biochem.J., 456, 2013
|
|
4ADZ
 
 | |
4AFM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens. | | Descriptor: | ACETATE ION, ENDOGLUCANASE CEL5A, GLYCEROL | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4BLF
 
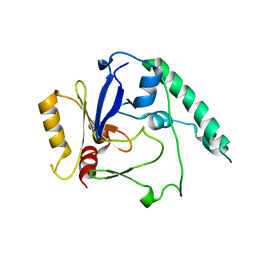 | | Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA. | | Descriptor: | SINGLE-STRAND DNA-BINDING PROTEIN | | Authors: | Bharat, T.A.M, Zbaida, D, Eisenstein, M, Frankenstein, Z, Mehlman, T, Weiner, L, Sorzano, C.O.S, Barak, Y, Albeck, S, Briggs, J.A.G, Wolf, S.G, Elbaum, M. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Variable Internal Flexibility Characterizes the Helical Capsid Formed by Agrobacterium Vire2 Protein on Single-Stranded DNA.
Structure, 21, 2013
|
|
4D1K
 
 | | Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. | | Descriptor: | GAG PROTEIN | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krauesslich, H.G, Briggs, J.A.G. | | Deposit date: | 2014-05-02 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CDU
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
3ZK0
 
 | |
3ZNJ
 
 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
4ARG
 
 | | Structure of the immature retroviral capsid at 8A resolution by cryo- electron microscopy | | Descriptor: | M-PMV DPRO CANC PROTEIN | | Authors: | Bharat, T.A.M, Davey, N.E, Ulbrich, P, Riches, J.D, Marco, A.D, Rumlova, M, Sachse, C, Ruml, T, Briggs, J.A.G. | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structure of the Immature Retroviral Capsid at 8A Resolution by Cryo-Electron Microscopy.
Nature, 487, 2012
|
|
4COC
 
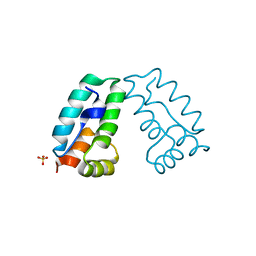 | | HIV-1 capsid C-terminal domain mutant (Y169L) | | Descriptor: | CAPSID PROTEIN P24, SULFATE ION | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4COP
 
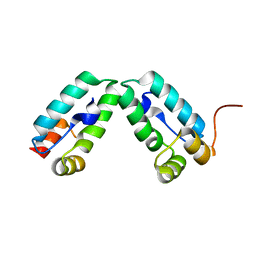 | | HIV-1 capsid C-terminal domain mutant (Y169S) | | Descriptor: | CAPSID PROTEIN P24 | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-29 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BZI
 
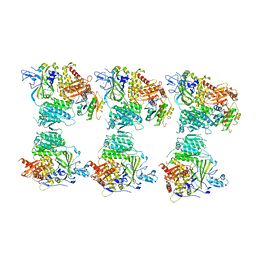 | | The structure of the COPII coat assembled on membranes | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SAR1P, ... | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4BZK
 
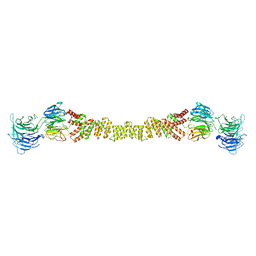 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
