1NK2
 
 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1XYS
 
 | | CATALYTIC CORE OF XYLANASE A E246C MUTANT | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1994-09-02 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the catalytic core of the family F xylanase from Pseudomonas fluorescens and identification of the xylopentaose-binding sites.
Structure, 2, 1994
|
|
1XOC
 
 | | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. | | Descriptor: | Nonapeptide VDSKNTSSW, Oligopeptide-binding protein appA, ZINC ION | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wright, L, Vagin, A.A, Wilkinson, A.J. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide.
J.Mol.Biol., 345, 2005
|
|
2UYW
 
 | | Crystal structure of Xenavidin | | Descriptor: | BIOTIN, FORMIC ACID, XENAVIDIN | | Authors: | Helppolainen, S.H, Maatta, J.A.E, Airenne, T.T, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-04-20 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characteristics of Xenavidin, the First Frog Avidin from Xenopus Tropicalis.
Bmc Struct.Biol., 9, 2009
|
|
4HDV
 
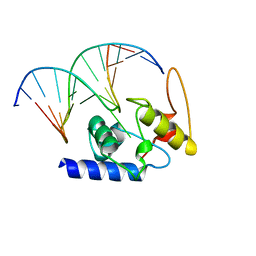 | |
1BZK
 
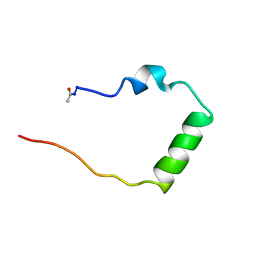 | | STRUCTURAL STUDIES ON THE EFFECTS OF THE DELETION IN THE RED CELL ANION EXCHANGER (BAND3, AE1) ASSOCIATED WITH SOUTH EAST ASIAN OVALOCYTOSIS. | | Descriptor: | PROTEIN (BAND 3 ANION TRANSPORT PROTEIN) | | Authors: | Chambers, E.J, Bloomberg, G.B, Ring, S.M, Tanner, M.J.A. | | Deposit date: | 1998-11-01 | | Release date: | 1999-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the effects of the deletion in the red cell anion exchanger (band 3, AE1) associated with South East Asian ovalocytosis.
J.Mol.Biol., 285, 1999
|
|
5T65
 
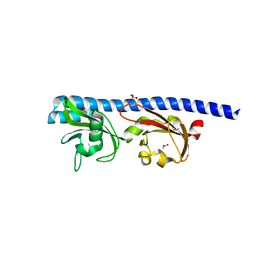 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-ILE | | Descriptor: | ACETATE ION, ISOLEUCINE, Methyl-accepting chemotaxis protein PctA, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
1XA8
 
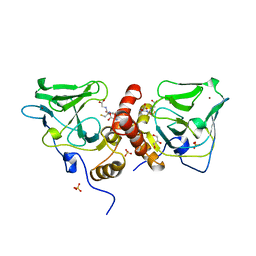 | | Crystal Structure Analysis of Glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, ... | | Authors: | Neculai, A.M, Neculai, D, Griesinger, C, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
5IGN
 
 | | Crystal structure of human BRD9 bromodomain in complex with LP99 chemical probe | | Descriptor: | Bromodomain-containing protein 9, N-[(2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl]-2-methylpropane-1-sulfonamide | | Authors: | Tallant, C, Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with LP99 chemical probe
To Be Published
|
|
1X6M
 
 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
2VN5
 
 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
1YAI
 
 | | X-RAY STRUCTURE OF A BACTERIAL COPPER,ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER, ZINC SUPEROXIDE DISMUTASE, ... | | Authors: | Bourne, Y, Redford, S.M, Lo, T.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 1996-02-03 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel dimeric interface and electrostatic recognition in bacterial Cu,Zn superoxide dismutase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1BTQ
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-11-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1YHG
 
 | |
2WAO
 
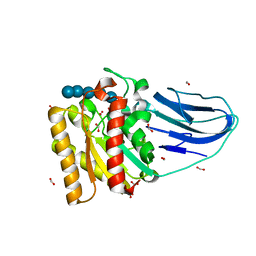 | | Structure of a family two carbohydrate esterase from Clostridium thermocellum in complex with cellohexaose | | Descriptor: | ENDOGLUCANASE E, FORMIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-10 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
1BH7
 
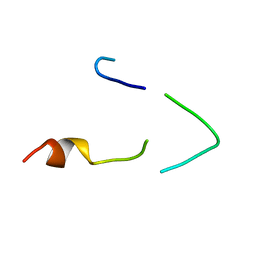 | | A LOW ENERGY STRUCTURE FOR THE FINAL CYTOPLASMIC LOOP OF BAND 3, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BAND 3 | | Authors: | Askin, D, Bloomberg, G.B, Chambers, E.J, Tanner, M.J.A. | | Deposit date: | 1998-06-16 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a cytoplasmic surface loop of the human red cell anion transporter, band 3.
Biochemistry, 37, 1998
|
|
2QFF
 
 | | Crystal structure of Staphylococcal Complement Inhibitor | | Descriptor: | Hypothetical protein | | Authors: | Milder, F.J, Rooijakkers, S.H.M, Bardoel, B.W, Ruyken, M, Van Strijp, J.A.G, Gros, P. | | Deposit date: | 2007-06-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Staphylococcal complement inhibitor: structure and active sites.
J.Immunol., 179, 2007
|
|
1BRV
 
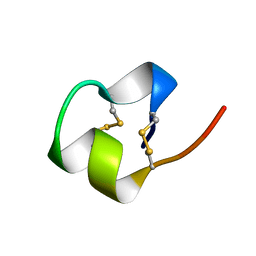 | | SOLUTION NMR STRUCTURE OF THE IMMUNODOMINANT REGION OF PROTEIN G OF BOVINE RESPIRATORY SYNCYTIAL VIRUS, 48 STRUCTURES | | Descriptor: | PROTEIN G | | Authors: | Doreleijers, J.F, Langedijk, J.P.M, Hard, K, Rullmann, J.A.C, Boelens, R, Schaaper, W.M, Van Oirschot, J.T, Kaptein, R. | | Deposit date: | 1996-03-29 | | Release date: | 1997-06-05 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant region of protein G of bovine respiratory syncytial virus.
Biochemistry, 35, 1996
|
|
1YHH
 
 | |
5T7M
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-TRP | | Descriptor: | ACETATE ION, Chemotaxis protein, SODIUM ION, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
1CCN
 
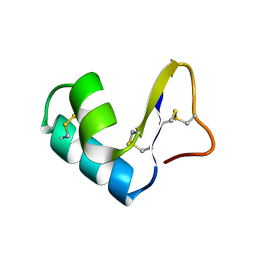 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Direct NOE refinement of biomolecular structures using 2D NMR data
J.Biomol.NMR, 1, 1991
|
|
1CCM
 
 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | "Ensemble" iterative relaxation matrix approach: a new NMR refinement protocol applied to the solution structure of crambin.
Proteins, 15, 1993
|
|
1BNX
 
 | | STRUCTURAL STUDIES ON THE EFFECTS OF THE DELETION IN THE RED CELL ANION EXCHANGER (BAND3, AE1) ASSOCIATED WITH SOUTH EAST ASIAN OVALOCYTOSIS. | | Descriptor: | PROTEIN (BAND 3) | | Authors: | Chambers, E.J, Bloomberg, G.B, Ring, S.M, Tanner, M.J.A. | | Deposit date: | 1998-07-30 | | Release date: | 1998-08-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Studies on the structure of a transmembrane region and a cytoplasmic loop of the human red cell anion exchanger (band 3, AE1).
Biochem.Soc.Trans., 26, 1998
|
|
2WAA
 
 | | Structure of a family two carbohydrate esterase from Cellvibrio japonicus | | Descriptor: | ACETATE ION, GLYCEROL, XYLAN ESTERASE, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2W5F
 
 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2008-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
