8U50
 
 | |
7BAY
 
 | |
8UIW
 
 | | yjdF riboswitch from R. gauvreauii in complex with chelerythrine bound to Fab BL3-6 S97N | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, ... | | Authors: | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | Deposit date: | 2023-10-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
8UTA
 
 | | yjdF riboswitch from R. gauvreauii in complex with proflavine bound to Fab BL3-6 S97N | | Descriptor: | Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, MAGNESIUM ION, ... | | Authors: | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
8U51
 
 | |
8U4Z
 
 | |
8Q4E
 
 | |
8QK8
 
 | |
1CPT
 
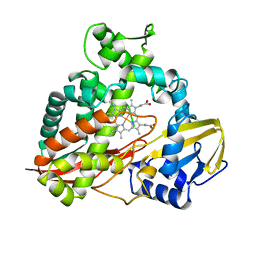 | | CRYSTAL STRUCTURE AND REFINEMENT OF CYTOCHROME P450-TERP AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME P450-TERP, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasemann, C.A, Ravichandran, K.G, Peterson, J.A, Deisenhofer, J. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and refinement of cytochrome P450terp at 2.3 A resolution.
J.Mol.Biol., 236, 1994
|
|
7BGS
 
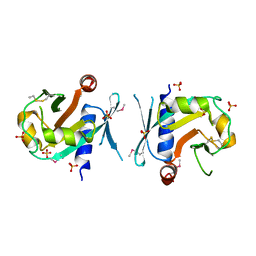 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BNX
 
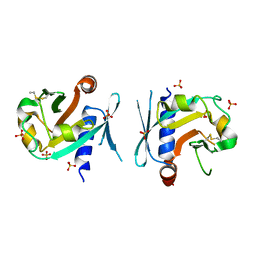 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BKW
 
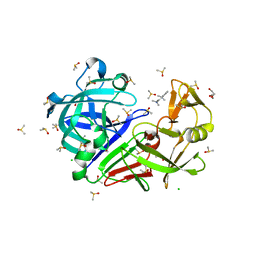 | | Endothiapepsin structure obtained at 100K with fragment BTB09871 bound | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Endothiapepsin structure obtained at 100K with fragment BTB09871 bound
To Be Published
|
|
8QIC
 
 | |
7BKR
 
 | | Endothiapepsin structure obtained at 298K and 40 mM DMSO from a dataset collected with JUNGFRAU detector | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endothiapepsin structure obtained at 298K and 40 mM DMSO from a dataset collected with JUNGFRAU detector
To Be Published
|
|
7BKV
 
 | | Endothiapepsin structure obtained at 100K with fragment AC39729 bound | | Descriptor: | 5-fluoranylpyridin-2-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Endothiapepsin structure obtained at 100K with fragment AC39729 bound
To Be Published
|
|
7BKY
 
 | | Endothiapepsin structure obtained at 298K with fragment BTB09871 bound from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, PENTAETHYLENE GLYCOL, ... | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Endothiapepsin structure obtained at 298K with fragment BTB09871 bound from a dataset collected with JUNGFRAU detector
To Be Published
|
|
8QME
 
 | |
7BKS
 
 | | 100K endothiapepsin structure obtained in presence of 40 mM DMSO | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | 100K endothiapepsin structure obtained in presence of 40 mM DMSO
To Be Published
|
|
7BKZ
 
 | | Endothiapepsin structure obtained at 298K after a soaking with fragment AC39729 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment AC39729 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
7BKU
 
 | | Endothiapepsin structure obtained at 100K with fragment JFD03909 bound | | Descriptor: | 1,10-PHENANTHROLINE, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Endothiapepsin structure obtained at 100K with fragment JFD03909 bound
To Be Published
|
|
8Q52
 
 | | A PBP-like protein built from fragments of different folds | | Descriptor: | Leucine-specific-binding protein,Chemotaxis protein CheY, SULFATE ION | | Authors: | Shanmugaratnam, S, Toledo-Patino, S, Goetz, S.K, Farias-Rico, J.A, Hocker, B. | | Deposit date: | 2023-08-08 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular handcraft of a well-folded protein chimera.
Febs Lett., 598, 2024
|
|
8Q3L
 
 | |
8QQ5
 
 | | Structure of WT SpNox DH domain: a bacterial NADPH oxidase. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Thepaut, M, Petit-Hartlein, I, Vermot, A, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
8QQ1
 
 | | SpNOX dehydrogenase domain, mutant F397W in complex with Flavin adenine dinucleotide (FAD) | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Humm, A.S, Dupeux, F, Vermot, A, Petit-Harleim, I, Fieschi, F, Marquez, J.A. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
8QQ7
 
 | | Structure of SpNOX: a Bacterial NADPH oxidase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-binding FR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Thepaut, M, Petit-Hartlein, I, Vermot, A, Chaptal, V, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
