5RMF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z54226006 | | Descriptor: | (2,6-difluorophenyl)(pyrrolidin-1-yl)methanone, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RYT
 
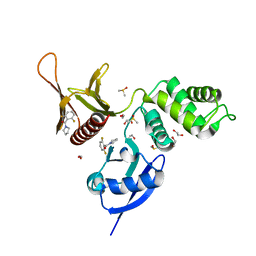 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z54226006 | | Descriptor: | (2,6-difluorophenyl)(pyrrolidin-1-yl)methanone, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZ5
 
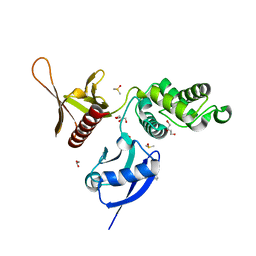 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434851 | | Descriptor: | 1,2-ETHANEDIOL, 4-({[(4-fluorophenyl)methyl]amino}methyl)benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZG
 
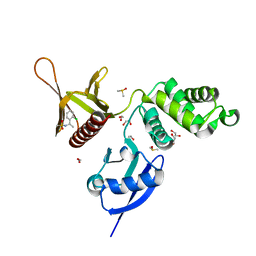 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z133729708 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZW
 
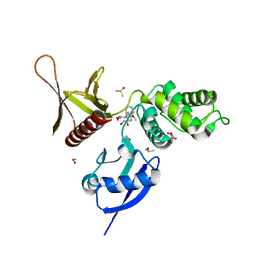 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2574937229 | | Descriptor: | (S)-1-(4-methoxyphenyl)-1-phenylmethanamine, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZ6
 
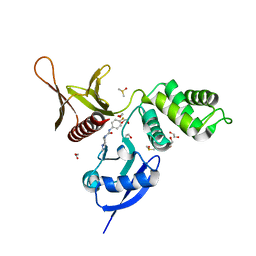 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434944 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZM
 
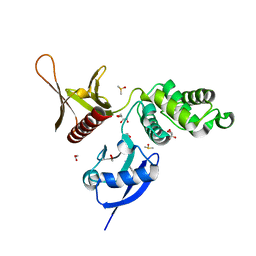 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434815 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(dimethylamino)methyl]benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5ROB
 
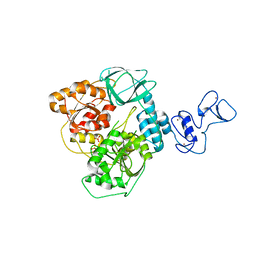 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 helicase | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
1IKG
 
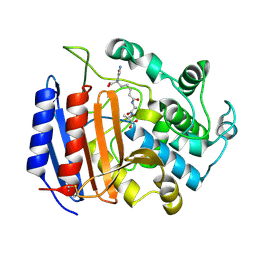 | | MICHAELIS COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
1O9A
 
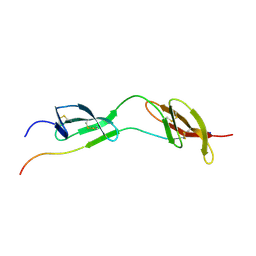 | | Solution structure of the complex of 1F12F1 from fibronectin with B3 from FnBB from S. dysgalactiae | | Descriptor: | FIBRONECTIN, FIBRONECTIN BINDING PROTEIN | | Authors: | Schwarz-Linek, U, Werner, J.M, Pickford, A.R, Pilka, E.S, Gurusiddappa, S, Briggs, J.A.G, Hook, M, Campbell, I.D, Potts, J.R. | | Deposit date: | 2002-12-11 | | Release date: | 2003-05-08 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper.
Nature, 423, 2003
|
|
3N3A
 
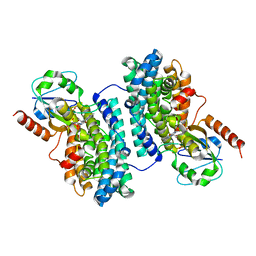 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli in Complex with Reduced NrdI | | Descriptor: | FLAVIN MONONUCLEOTIDE, MANGANESE (II) ION, Protein nrdI, ... | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
3N39
 
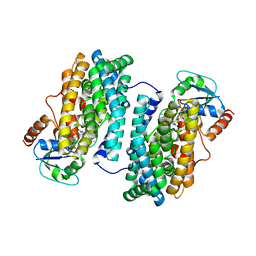 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli in Complex with NrdI | | Descriptor: | FLAVIN MONONUCLEOTIDE, MANGANESE (II) ION, Protein nrdI, ... | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
5UCF
 
 | | Solution NMR-derived model of the minor species of DANCER-2, a dynamic and natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
5UB0
 
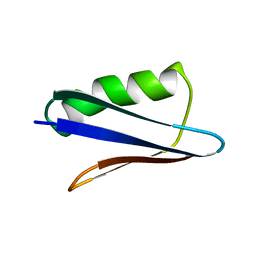 | | Solution NMR Structure of NERD-C, a natively folded tetramutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
5UIB
 
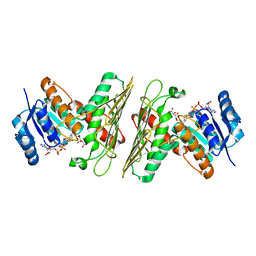 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, L-tartaric acid and Magnesium | | Descriptor: | L(+)-TARTARIC ACID, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, L-tartaric acid and Magnesium
To be published
|
|
5UIA
 
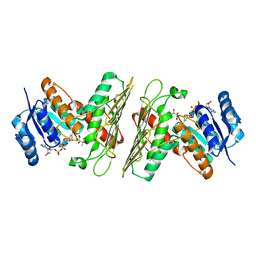 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, R-2,3-dihydroxyisovalerate and Magnesium | | Descriptor: | (2S)-2,3-dihydroxy-3-methylbutanoic acid, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, R-2,3-dihydroxyisovalerate and Magnesium
To be published
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5UOO
 
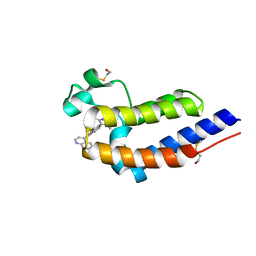 | | BRD4 bromodomain 2 in complex with CD161 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indole, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-05-17 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Based Discovery of 4-(6-Methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indol-7-yl)-3,5-dimethylisoxazole (CD161) as a Potent and Orally Bioavailable BET Bromodomain Inhibitor.
J. Med. Chem., 60, 2017
|
|
5U8U
 
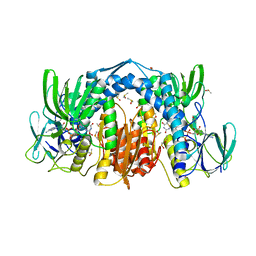 | | Dihydrolipoamide dehydrogenase (LpdG) from Pseudomonas aeruginosa | | Descriptor: | DIMETHYL SULFOXIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Glasser, N.R, Wang, B.X, Hoy, J.A, Newman, D.K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Pyruvate and alpha-Ketoglutarate Dehydrogenase Complexes of Pseudomonas aeruginosa Catalyze Pyocyanin and Phenazine-1-carboxylic Acid Reduction via the Subunit Dihydrolipoamide Dehydrogenase.
J. Biol. Chem., 292, 2017
|
|
3N5F
 
 | | Crystal Structure of L-N-carbamoylase from Geobacillus stearothermophilus CECT43 | | Descriptor: | CACODYLATE ION, COBALT (II) ION, ISOPROPYL ALCOHOL, ... | | Authors: | Garcia-Pino, A, Martinez-Rodriguez, S, Gavira, J.A. | | Deposit date: | 2010-05-25 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mutational and structural analysis of L-N-carbamoylase reveals new insights into a peptidase m20/m25/m40 family member.
J.Bacteriol., 194, 2012
|
|
5U9T
 
 | | The Tris-thiolate Zn(II)S3Cl Binding Site Engineered by D-Cysteine Ligands in de Novo Three-stranded Coiled Coil Environment | | Descriptor: | ACETATE ION, CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Ruckthong, L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | d-Cysteine Ligands Control Metal Geometries within De Novo Designed Three-Stranded Coiled Coils.
Chemistry, 23, 2017
|
|
1NWW
 
 | | Limonene-1,2-epoxide hydrolase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEPTANAMIDE, Limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
5UFQ
 
 | | K-RasG12D(GNP)/R11.1.6 complex | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, J.A, Mattos, C. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | An engineered protein antagonist of K-Ras/B-Raf interaction.
Sci Rep, 7, 2017
|
|
5UHW
 
 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+ and Magnesium | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase protein | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+ and Magnesium
To be published
|
|
5UHZ
 
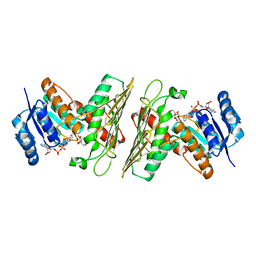 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, D-Apionate and Magnesium | | Descriptor: | (3R,4R)-3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-one, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, D-Apionate and Magnesium
To be published
|
|
