6VYP
 
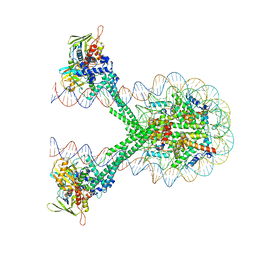 | | Crystal structure of the LSD1/CoREST histone demethylase bound to its nucleosome substrate | | Descriptor: | DNA (191-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A type 1, ... | | Authors: | Kim, S, Zhu, J, Eek, P, Yennawar, N, Song, T. | | Deposit date: | 2020-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.99 Å) | | Cite: | Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate.
Mol.Cell, 78, 2020
|
|
6PH3
 
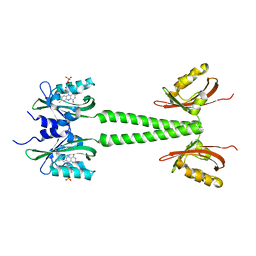 | | LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (dark-adapted, construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
7QSX
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8 complex | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
3PB3
 
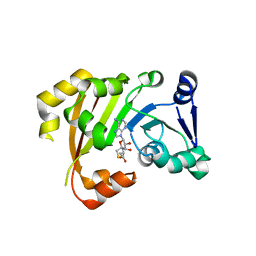 | | Structure of an Antibiotic Related Methyltransferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit.
Nucleic Acids Res., 2010
|
|
8TDK
 
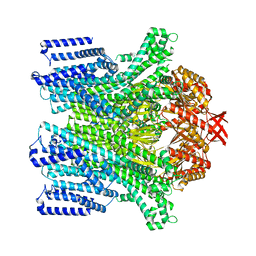 | | Cryo-EM structure of AtMSL10-G556V | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDL
 
 | |
7QY5
 
 | | Crystal structure of the S.pombe Ars2-Red1 complex. | | Descriptor: | NURS complex subunit pir2, RNA elimination defective protein Red1, ZINC ION | | Authors: | Foucher, A.E, Kadlec, J. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
8SSD
 
 | |
1JB1
 
 | | Lactobacillus casei HprK/P Bound to Phosphate | | Descriptor: | HPRK PROTEIN, PHOSPHATE ION | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Monedero, V, Gueguen-Chaignon, V, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain.
EMBO J., 20, 2001
|
|
8SSE
 
 | | Methionine synthase, C-terminal fragment, Cobalamin and Reactivation Domains from Thermus thermophilus HB8 | | Descriptor: | COBALAMIN, Methionine synthase | | Authors: | Yamada, K, Mendoza, J, Koutmos, M. | | Deposit date: | 2023-05-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of full-length cobalamin-dependent methionine synthase and cofactor loading captured in crystallo.
Nat Commun, 14, 2023
|
|
6PKL
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.59A resolution (#7) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.59 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
7FB5
 
 | | Crystal structure of FAM134B/GABARAP complex | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Reticulophagy regulator 1 | | Authors: | Zhao, J, Li, J. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The crystal structure of the FAM134B-GABARAP complex provides mechanistic insights into the selective binding of FAM134 to the GABARAP subfamily.
Febs Open Bio, 12, 2022
|
|
6PKT
 
 | | MicroED structure of proteinase K from merging low-dose, platinum pre-coated lamellae at 1.85A resolution (LDPT) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
5YJG
 
 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
2WID
 
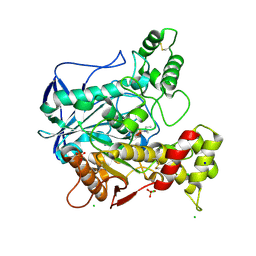 | | NONAGED FORM OF HUMAN BUTYRYLCHOLINESTERASE INHIBITED BY TABUN ANALOGUE TA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Carletti, E, Aurbek, N, Gillon, E, Loiodice, M, Nicolet, Y, Fontecilla, J, Masson, P, Thiermann, H, Nachon, F, Worek, F. | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity analysis of aging and reactivation of human butyrylcholinesterase inhibited by analogues of tabun.
Biochem. J., 421, 2009
|
|
6VRI
 
 | | Crystal Structure of the wtBlc-split Protein | | Descriptor: | Outer membrane lipoprotein Blc, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Bozhanova, N.G, Meiler, J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lipocalin Blc is a potential heme-binding protein.
Febs Lett., 595, 2021
|
|
2K5Q
 
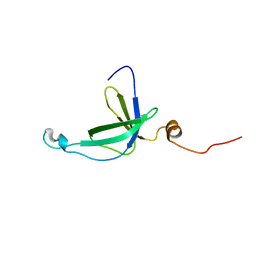 | | NMR Solution structure of membrane associated protein from Bacillus cereus: Northeast Structural Genomics Consortium Target: BcR97A | | Descriptor: | Hypothetical Membrane Associated Protein BcR97A | | Authors: | Swapna, G, Wang, D, Owens, L, Xiao, R, Liu, J, Baran, M.C, Everett, J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution Structure of Membrane associated protein from Bacillus cereus: Northeast Structural Genomics Consortium Target: BcR97A
To be Published
|
|
6PPB
 
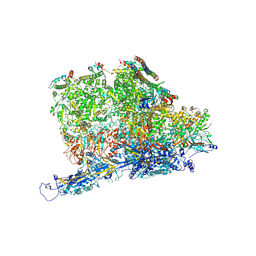 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C5 portal vertex structure | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
8SUZ
 
 | | Open State of the SARS-CoV-2 Envelope Protein Transmembrane Domain, Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Medeiros-Silva, J, Dregni, A.J, Somberg, N.H, Hong, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure of the open SARS-CoV-2 E viroporin.
Sci Adv, 9, 2023
|
|
2WIF
 
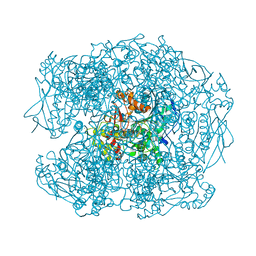 | | AGED FORM OF HUMAN BUTYRYLCHOLINESTERASE INHIBITED BY TABUN ANALOGUE TA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Carletti, E, Aurbek, N, Gillon, E, Loiodice, M, Nicolet, Y, Fontecilla, J, Masson, P, Thiermann, H, Nachon, F, Worek, F. | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-activity analysis of aging and reactivation of human butyrylcholinesterase inhibited by analogues of tabun.
Biochem. J., 421, 2009
|
|
7QVM
 
 | | Human Oxytocin receptor (OTR) oxytocin Gq chimera (mGoqi) complex | | Descriptor: | Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Waltenspuhl, Y, Ehrenmann, J, Vacca, S, Thom, C, Medalia, O, Pluckthun, A. | | Deposit date: | 2022-01-21 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for the activation and ligand recognition of the human oxytocin receptor.
Nat Commun, 13, 2022
|
|
7XHQ
 
 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[(3S,4R)-4-azanyl-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-04-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the SHP2 allosteric inhibitor 2-((3R,4R)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl)-5-(2,3-dichlorophenyl)-3-methylpyrrolo[2,1-f][1,2,4] triazin-4(3H)-one.
J Enzyme Inhib Med Chem, 38, 2023
|
|
6W4X
 
 | | Holocomplex of E. coli class Ia ribonucleotide reductase with GDP and TTP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MU-OXO-DIIRON, ... | | Authors: | Kang, G, Taguchi, A, Stubbe, J, Drennan, C. | | Deposit date: | 2020-03-11 | | Release date: | 2020-04-08 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex.
Science, 368, 2020
|
|
6W69
 
 | | The structure of F64, S172A Keap1-BTB domain | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6W6D
 
 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
