8A65
 
 | | Small molecule stabilizer (compound 3) for FOXO1 and 14-3-3 | | Descriptor: | (3~{S})-1-[2-azanyl-3,5-bis(chloranyl)phenyl]carbonyl-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-3-carboxamide, 14-3-3 protein sigma, Forkhead box protein O1, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
6OC6
 
 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum and Pyrroloquinoline quinone | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
8A68
 
 | | Small molecule stabilizer (compound 5) for C-RAF(pS259) and 14-3-3 | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, RAF proto-oncogene serine/threonine-protein kinase, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
6UVY
 
 | | BACE-1 in complex with compound #18 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
7PQR
 
 | | LsAA9A expressed in E. coli | | Descriptor: | ACETATE ION, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Muderspach, S.J, Metherall, J, Ipsen, J, Rollan, C.H, Norholm, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
8A6F
 
 | | Small molecule stabilizer (compound 8) for C-RAF and 14-3-3 | | Descriptor: | 1-[4-[4-chloranyl-3-(trifluoromethyl)phenyl]-4-oxidanyl-piperidin-1-yl]-3-[2-(dimethylamino)ethyldisulfanyl]propan-1-one, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
6UZZ
 
 | | structure of human KCNQ1-CaM complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mackinnon, R, Sun, J. | | Deposit date: | 2019-11-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Human KCNQ1 Modulation and Gating.
Cell, 180, 2020
|
|
2MEQ
 
 | |
5XWV
 
 | |
2WU7
 
 | | Crystal Structure of the Human CLK3 in complex with V25 | | Descriptor: | CHLORIDE ION, DUAL SPECIFICITY PROTEIN KINASE CLK3, SULFATE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Phillips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Bracher, F, Huber, K, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
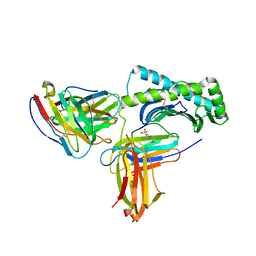
6UZ1
 
 | |
6V2S
 
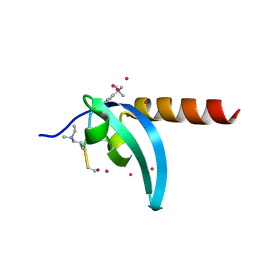 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | Descriptor: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
7ZQU
 
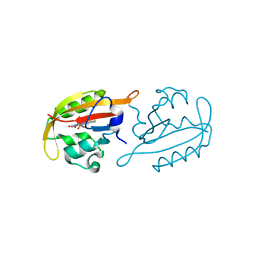 | |
7ZYN
 
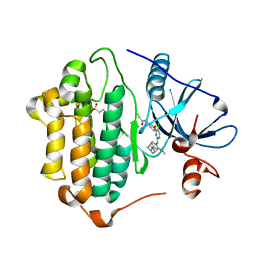 | | Crystal Structure of EGFR-T790M/C797S in Complex with WZ4002 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, N-{3-[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
7PFL
 
 | | The SARS-CoV2 major protease (Mpro) apo structure to 1.8 A resolution | | Descriptor: | Replicase polyprotein 1ab, SODIUM ION | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
2X12
 
 | |
1BGA
 
 | |
6OF9
 
 | |
7ZYM
 
 | | Crystal Structure of EGFR-T790M/C797S in Complex with Brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
6V0Y
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 72,74cit69-81, GLYCEROL, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
7ZYP
 
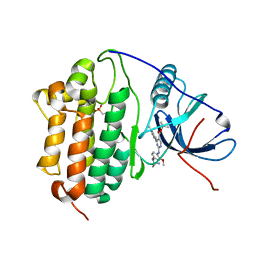 | | Crystal Structure of EGFR-T790M/C797S in Complex with Reversible Aminopyrimidine 9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, propan-2-yl 2-[[4-(4-azanylpiperidin-1-yl)-2-methoxy-phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
6OPB
 
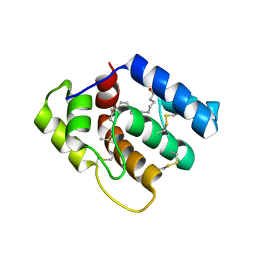 | |
6OPK
 
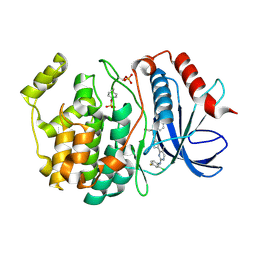 | | Phosphorylated ERK2 with Vertex-11e | | Descriptor: | 4-{2-[(2-chloro-4-fluorophenyl)amino]-5-methylpyrimidin-4-yl}-N-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-1H-pyrrole-2-carboxamide, Mitogen-activated protein kinase 1 | | Authors: | Vigers, G.P, Rudolph, J. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | Descriptor: | CALCIUM ION, hydrolase Ple628 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7PDY
 
 | | A viral peptide from Marek's disease virus bound to chicken MHC-II molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 38 kDa phosphoprotein,MHC class II beta chain, ... | | Authors: | Goryanin, A, Cook, A.G, Kaufman, J, Halabi, S. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Viral peptide bound to chicken MHC-II molecule
To Be Published
|
|
