3L3J
 
 | | Crystal structure of HLA-B*4402 in complex with the F3A/R5A double mutant of a self-peptide derived from DPA*0201 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3L3U
 
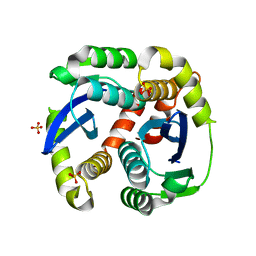 | | Crystal structure of the HIV-1 integrase core domain to 1.4A | | Descriptor: | POL polyprotein, SULFATE ION | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site.
Febs Lett., 584, 2010
|
|
4AFU
 
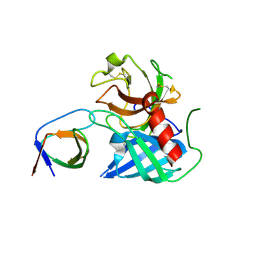 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
6CUS
 
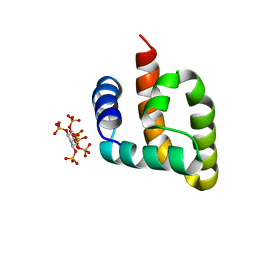 | |
2NSK
 
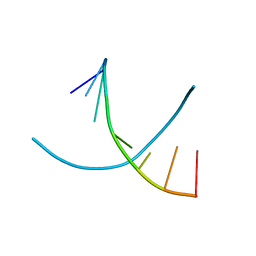 | | Doubled Modified Selenium DNA | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T4S)P*AP*CP*AP*C)-3' | | Authors: | Jiang, J, Shen, J, Huang, Z. | | Deposit date: | 2006-11-04 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Double Modified Selinium DNA [G(Use)G(Tse)ACAC]
To be Published
|
|
7OXI
 
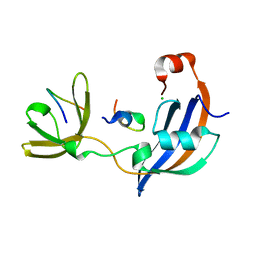 | | ttSlyD with W4A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXK
 
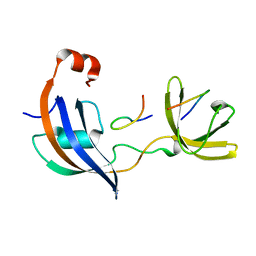 | | ttSlyD with W4K pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, Peptidyl-prolyl cis-trans isomerase | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXJ
 
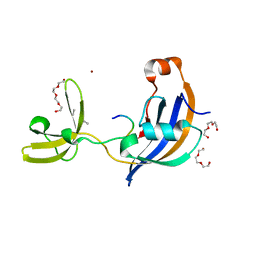 | | ttSlyD with M8A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, Fragment of 30S ribosomal protein S2 peptide, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXG
 
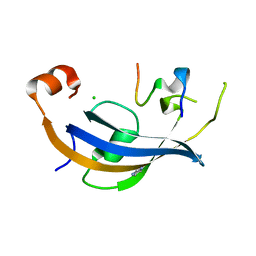 | | ttSlyD FKBP domain with M8A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXH
 
 | | ttSlyD with pseudo-wild-type S2 peptide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 30S ribosomal protein S2, CHLORIDE ION, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
4AP7
 
 | | Crystal structure of C-MET kinase domain in complex with 4-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl)phenol | | Descriptor: | 4-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]phenol, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
7OUO
 
 | | Crystal structure of RNA duplex [UCGUGCGA]2 in complex with Ba2+ cation | | Descriptor: | BARIUM ION, RNA (5'-R(*UP*CP*GP*UP*GP*CP*GP*A)-3') | | Authors: | Ruszkowski, M, Mao, S, Zheng, Y.Y, Ruszkowska, A, Sheng, J. | | Deposit date: | 2021-06-12 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Structural Insights Into the 5'UG/3'GU Wobble Tandem in Complex With Ba 2+ Cation.
Front Mol Biosci, 8, 2021
|
|
6CUP
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | FORMIC ACID, GLYCEROL, GTPase HRas, ... | | Authors: | Phan, J, Abbott, J, Fesik, S.W. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Discovery of Quinazolines That Activate SOS1-Mediated Nucleotide Exchange on RAS.
ACS Med Chem Lett, 9, 2018
|
|
2GV0
 
 | |
4AQH
 
 | | Plasminogen activator inhibitor type-1 in complex with the inhibitor AZ3976 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR 1, TERT-BUTYL 3-[(4-OXO-3H-PYRIDO[2,3-D]PYRIMIDIN-2-YL)AMINO]AZETIDINE-1-CARBOXYLATE | | Authors: | Fjellstrom, O, Deinum, J, Sjogren, T, Johansson, C, Geschwindner, S, Nerme, V, Legnehed, A, McPheat, J, Olsson, K, Bodin, C, Gustafsson, D. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a Small Molecule Inhibitor of Plasminogen Activator Inhibitor Type 1 that Accelerates the Transition Into the Latent Conformation
J.Biol.Chem., 288, 2013
|
|
2NY0
 
 | | HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, T257S, V275C, S334A, S375W, A433M) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
4A6J
 
 | | Structural model of ParM filament based on CryoEM map | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Gayathri, P, Fujii, T, Moller-Jensen, J, Van Den Ent, F, Namba, K, Lowe, J. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
3L3D
 
 | | Crystal structure of HLA-B*4402 in complex with the F3A mutant of a self-peptide derived from DPA*0201 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2O1Z
 
 | | Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155) | | Descriptor: | FE (III) ION, Ribonucleotide Reductase Subunit R2, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Tempel, W, Qiu, W, Lew, J, Wernimont, A.K, Lin, Y.H, Hassanali, A, Melone, M, Zhao, Y, Nordlund, P, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155)
To be Published
|
|
4A9G
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
6BHG
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
3DG0
 
 | |
4ABY
 
 | | Crystal structure of Deinococcus radiodurans RecN head domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
3DH1
 
 | | Crystal structure of human tRNA-specific adenosine-34 deaminase subunit ADAT2 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase 2 | | Authors: | Welin, M, Tresaugues, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, van der Berg, S, Wisniewska, M, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human tRNA-specific adenosine-34 deaminase subunit ADAT2.
To be Published
|
|
3L3K
 
 | | Crystal structure of HLA-B*4402 in complex with the R5A/F7A double mutant of a self-peptide derived from DPA*0201 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
