6HFG
 
 | |
6HG6
 
 | | Clostridium beijerinckii aldo-keto reductase Cbei_3974 with NADPH | | Descriptor: | 1,2-ETHANEDIOL, L-glyceraldehyde 3-phosphate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scott, A.F, Cresser-Brown, J, Rizkallah, P.J, Jin, Y, Allemann, R.K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Biophysical Analysis of Furfural-Detoxifying Aldehyde Reductase from Clostridium beijerinckii.
Appl.Environ.Microbiol., 85, 2019
|
|
5QI7
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000506a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
2VWL
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3R,5S)-1-{[2-FLUORO-4-(2-OXO-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-5-HYDROXYMETHYL-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VXR
 
 | |
1HI7
 
 | |
2W9E
 
 | | Structure of ICSM 18 (anti-Prp therapeutic antibody) Fab fragment complexed with human Prp fragment 119-231 | | Descriptor: | ICSM 18-ANTI-PRP THERAPEUTIC FAB HEAVY CHAIN, ICSM 18-ANTI-PRP THERAPEUTIC FAB LIGHT CHAIN, MAJOR PRION PROTEIN, ... | | Authors: | Antonyuk, S.V, Trevitt, C.R, Strange, R.W, Jackson, G.S, Sangar, D, Batchelor, M, Jones, S, Georgiou, T, Cooper, S, Fraser, C, Khalili-Shirazi, A, Clarke, A.R, Hasnain, S.S, Collinge, J. | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Prion Protein Bound to a Therapeutic Antibody.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W0R
 
 | | Crystal structure of the mutated N263D YscU C-terminal domain | | Descriptor: | CHLORIDE ION, YSCU | | Authors: | Wiesand, U, Sorg, I, Amstutz, M, Wagner, S, Van Den Heuvel, J, Luehrs, T, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the Type III Secretion Recognition Protein Yscu from Yersinia Enterocolitica
J.Mol.Biol., 385, 2009
|
|
5QHU
 
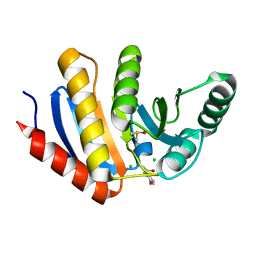 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMSOA000341b | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N-(2-hydroxyphenyl)acetamide, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
2VYV
 
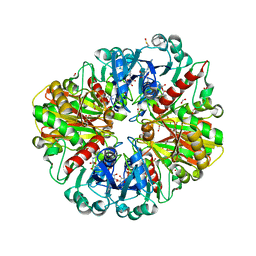 | | Structure of E.Coli GAPDH Rat Sperm GAPDH heterotetramer | | Descriptor: | FORMIC ACID, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Frayne, J, Taylor, A, Hall, L, Hadfield, A. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of Insoluble Rat Sperm Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) Via Heterotetramer Formation with Escherichia Coli Gapdh Reveals Target for Contraceptive Design.
J.Biol.Chem., 284, 2009
|
|
2WBD
 
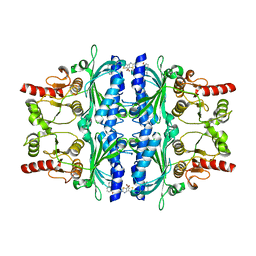 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-3-ethylbenzenesulfonamide | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5M54
 
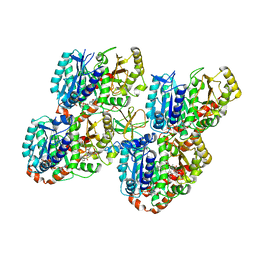 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-J, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2W9F
 
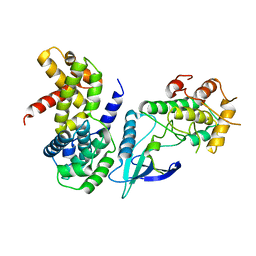 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-23 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6JPU
 
 | | CryoEM structure of Abo1 hexamer - apo complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
1HNH
 
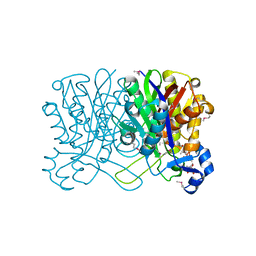 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III + DEGRADED FORM OF ACETYL-COA | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, COENZYME A | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Head, M, Lonsdale, J, Konstantinidis, A.K. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of beta-ketoacyl-acyl carrier protein synthase III.
J.Mol.Biol., 307, 2001
|
|
2VRO
 
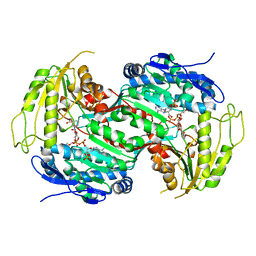 | | Crystal structure of aldehyde dehydrogenase from Burkholderia xenovorans LB400 | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALDEHYDE DEHYDROGENASE, HEXAETHYLENE GLYCOL, ... | | Authors: | Bains, J, Boulanger, M.J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of a Novel Aldehyde Dehydrogenase Encoded by the Benzoate Oxidation (Box) Pathway in Burkholderia Xenovorans Lb400
J.Mol.Biol., 379, 2008
|
|
2VUF
 
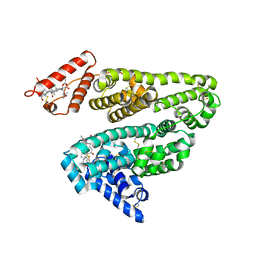 | |
1HND
 
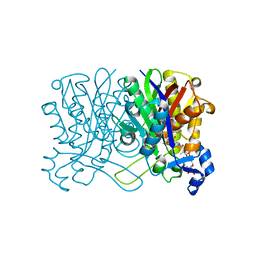 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III-COA COMPLEX | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, COENZYME A | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Head, M, Lonsdale, J, Konstantinidis, A.K. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined structures of beta-ketoacyl-acyl carrier protein synthase III.
J.Mol.Biol., 307, 2001
|
|
2VUU
 
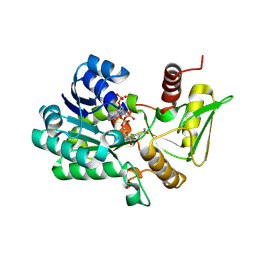 | | Crystal structure of NADP-bound NmrA-AreA zinc finger complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5MTA
 
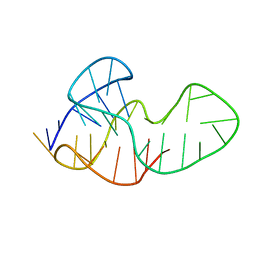 | |
2W78
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(ATCC13535)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
5MEE
 
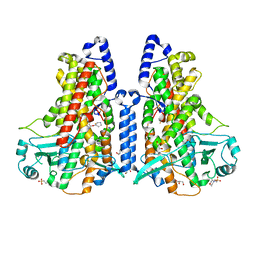 | | Cyanothece lipoxygenase 2 (CspLOX2) variant - L304V | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Arachidonate 15-lipoxygenase, FE (III) ION, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
2W3R
 
 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in complex with hypoxanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
5MUK
 
 | | Glycoside Hydrolase BT3686 | | Descriptor: | Neuraminidase | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
