4PBC
 
 | |
5FNF
 
 | | Dynamic Undocking and the Quasi-Bound State as tools for Drug Design | | Descriptor: | 4-[(E)-N-oxidanyl-C-pyridin-3-yl-carbonimidoyl]benzene-1,3-diol, HEAT SHOCK PROTEIN, HSP90-ALPHA, ... | | Authors: | Ruiz-Carmona, S, Schmidtke, P, Luque, F.J, Baker, L.M, Matassova, N, Davis, B, Roughley, S, Murray, J, Hubbard, R, Barril, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamic undocking and the quasi-bound state as tools for drug discovery.
Nat Chem, 9, 2017
|
|
5YNY
 
 | | Structure of house dust mite allergen Der F 21 in PEG2KMME | | Descriptor: | Allergen Der f 21 | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
4PMD
 
 | |
5FCI
 
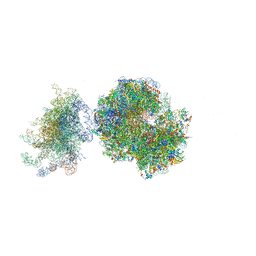 | | Structure of the vacant uL3 W255C mutant 80S yeast ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Mailliot, J, Garreau de Loubresse, N, Yusupova, G, Dinman, J.D, Yusupov, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structures of the uL3 Mutant Ribosome: Illustration of the Importance of Ribosomal Proteins for Translation Efficiency.
J.Mol.Biol., 428, 2016
|
|
5G0V
 
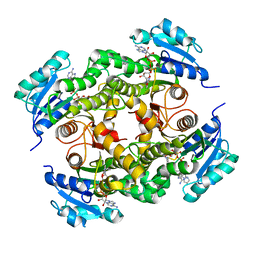 | | InhA in complex with a DNA encoded library hit | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN REDUCTASE, MAGNESIUM ION, N-[2-(methylamino)-2-oxidanylidene-ethyl]-2-(4-pyrazol-1-ylphenyl)-N-(1-pyridin-2-ylpiperidin-4-yl)ethanamide, ... | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Cofactor-Specific, Bactericidal Mycobacterium Tuberculosis Inha Inhibitors Using DNA-Encoded Library Technology
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4PNS
 
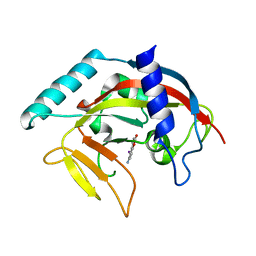 | | Crystal Structure of human Tankyrase 2 in complex with INH2BP. | | Descriptor: | 6-amino-5-iodo-2H-chromen-2-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8JL3
 
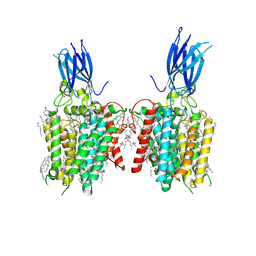 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | membrane proteins
To Be Published
|
|
5FS5
 
 | |
8J4T
 
 | | GajA-GajB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Wei, T, Ma, J, Huo, Y. | | Deposit date: | 2023-04-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and biochemical insights into the mechanism of the Gabija bacterial immunity system.
Nat Commun, 15, 2024
|
|
4PR6
 
 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
5FN7
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
1AJK
 
 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
8IVP
 
 | | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase, D-fructose | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii
To Be Published
|
|
8JL4
 
 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | membrane proteins
To Be Published
|
|
5DS0
 
 | | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09 | | Descriptor: | COBALT (II) ION, GLYCEROL, Peptidase M42 | | Authors: | Michalska, K, Chhor, G, Mootz, J, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09
To Be Published
|
|
5DUO
 
 | | Crystal structure of native translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FORMIC ACID, PROTOPORPHYRIN IX, ... | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2015-09-20 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response to Comment on "Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism".
Science, 350, 2015
|
|
1YE9
 
 | | Crystal structure of proteolytically truncated catalase HPII from E. coli | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, catalase HPII | | Authors: | Loewen, P.C, Chelikani, P, Carpena, X, Fita, I, Perez-Luque, R, Donald, L.J, Switala, J, Duckworth, H.W. | | Deposit date: | 2004-12-28 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of a Large Subunit Catalase Truncated by Proteolytic Cleavage(,)
Biochemistry, 44, 2005
|
|
8J0Y
 
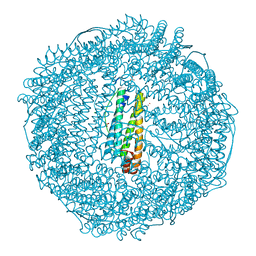 | | Crystal structure of horse spleen L-ferritin at 20deg Celsius. | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Level Insights into a Unique Semi-Clathrate Hydrate Formed in a Confined Environment of Porous Protein Crystal.
Cryst.Growth Des., 23, 2023
|
|
3P8H
 
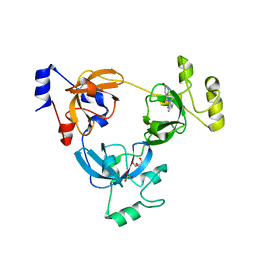 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
1XB7
 
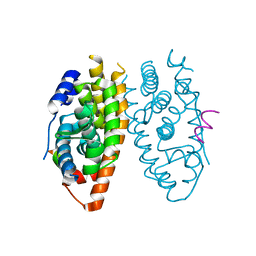 | | X-ray structure of ERRalpha LBD in complex with a PGC-1alpha peptide at 2.5A resolution | | Descriptor: | IODIDE ION, Peroxisome proliferator activated receptor gamma coactivator 1 alpha, Steroid hormone receptor ERR1 | | Authors: | Kallen, J, Schlaeppi, J.M, Bitsch, F, Filipuzzi, I, Schilb, A, Riou, V, Graham, A, Strauss, A, Geiser, M, Fournier, B. | | Deposit date: | 2004-08-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for Ligand-independent Transcriptional Activation of the Human Estrogen-related Receptor {alpha} (ERR{alpha}): CRYSTAL STRUCTURE OF ERR{alpha} LIGAND BINDING DOMAIN IN COMPLEX WITH PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR COACTIVATOR-1{alpha}
J.Biol.Chem., 279, 2004
|
|
1XGW
 
 | |
1XBZ
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound L-xylulose 5-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
8J0W
 
 | | Crystal structure of horse spleen L-ferritin at -40deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Level Insights into a Unique Semi-Clathrate Hydrate Formed in a Confined Environment of Porous Protein Crystal.
Cryst.Growth Des., 23, 2023
|
|
8J0Z
 
 | | Crystal structure of horse spleen L-ferritin at -180deg Celsius cooled from -40deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Level Insights into a Unique Semi-Clathrate Hydrate Formed in a Confined Environment of Porous Protein Crystal.
Cryst.Growth Des., 23, 2023
|
|
