2OCB
 
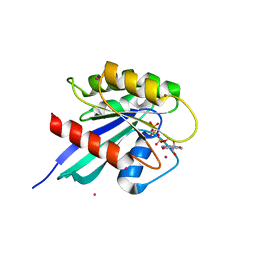 | | Crystal structure of human RAB9B in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-9B, ... | | Authors: | Hong, B, Shen, L, Walker, J.R, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human RAB9B in complex with a GTP analogue
To be Published
|
|
6KMZ
 
 | |
6CJT
 
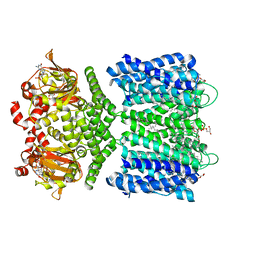 | | Structure of the SthK cyclic nucleotide-gated potassium channel in complex with cGMP | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CYCLIC GUANOSINE MONOPHOSPHATE, SthK cyclic nucleotide-gated potassium channel | | Authors: | Nimigean, C.M, Rheinberger, J. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures.
Elife, 7, 2018
|
|
6CLC
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.27 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
3IST
 
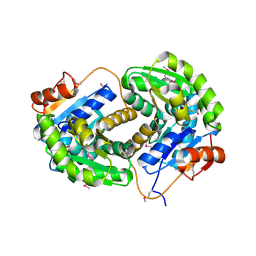 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid | | Descriptor: | CHLORIDE ION, Glutamate racemase, SUCCINIC ACID | | Authors: | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid
TO BE PUBLISHED
|
|
5TAU
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 3) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
3IT7
 
 | | Crystal Structure of the LasA virulence factor from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Protease lasA, ... | | Authors: | Spencer, J, Murphy, L.M, Conners, R, Sessions, R.B, Gamblin, S.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the LasA virulence factor from Pseudomonas aeruginosa: substrate specificity and mechanism of M23 metallopeptidases.
J.Mol.Biol., 396, 2010
|
|
4APC
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1 | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
6CLH
 
 | | 1.37 A MicroED structure of GSNQNNF at 2.9 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.37 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLP
 
 | | 1.16 A MicroED structure of GSNQNNF at 2.5 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.16 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
4AN3
 
 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
6K52
 
 | | Hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from the microbial flora of a Japanese hot spring | | Descriptor: | ACETATE ION, CALCIUM ION, GH6 cellobiohydrolase, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68000138 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
5TBF
 
 | |
4AFP
 
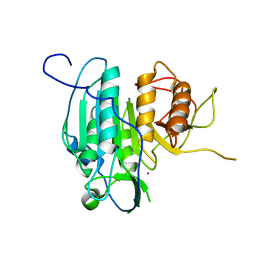 | | The structure of metacaspase 2 from T. brucei determined in the presence of Samarium | | Descriptor: | METACASPASE MCA2, SAMARIUM (III) ION | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A8X
 
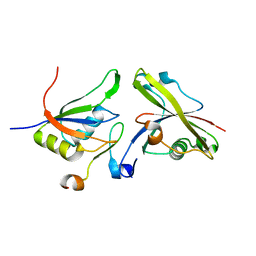 | | Structure of the core ASAP complex | | Descriptor: | HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, HOOK-LIKE, ISOFORM A, ... | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
3IV1
 
 | | Coiled-coil domain of tumor susceptibility gene 101 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor susceptibility gene 101 protein | | Authors: | Neculai, D, Avvakumov, G.V, Wernimont, A.K, Xue, S, Walker, J.R, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coiled-Coil Domain of Human Tsg101
To be Published
|
|
4AII
 
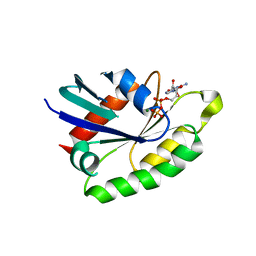 | | Crystal structure of the rat REM2 GTPase - G domain bound to GDP | | Descriptor: | GTP-BINDING PROTEIN REM 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Reymond, P, Coquard, A, Chenon, M, Zeghouf, M, El Marjou, A, Thompson, A, Menetrey, J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Gdp-Bound G Domain of the Rgk Protein Rem2.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5TBV
 
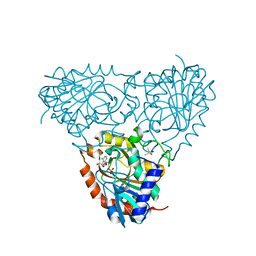 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase complexed with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase, ... | | Authors: | Faheem, M, Torini, J.R, Romanello, L, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase
To Be Published
|
|
4AOF
 
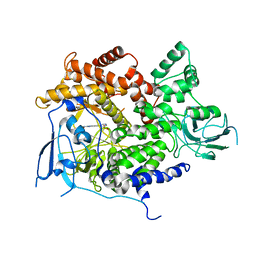 | | Selective small molecule inhibitor discovered by chemoproteomic assay platform reveals regulation of Th17 cell differentiation by PI3Kgamma | | Descriptor: | N-[6-(5-methylsulfonylpyridin-3-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]ethanamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Bergamini, G, Bell, K, Shimamura, S, Werner, T, Cansfield, A, Muller, K, Perrin, J, Rau, C, Ellard, K, Hopf, C, Doce, C, Leggate, D, Mangano, R, Mathieson, T, OMahony, A, Plavec, I, Rharbaoui, F, Reinhard, F, Savitski, M.M, Ramsden, N, Hirsch, E, Drewes, G, Rausch, O, Bantscheff, M, Neubauer, G. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Selective Inhibitor Reveals Pi3Kgamma Dependence of T(H)17 Cell Differentiation.
Nat.Chem.Biol., 8, 2012
|
|
5TDB
 
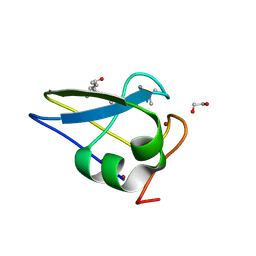 | | Crystal structure of the human UBR-box domain from UBR2 in complex with asymmetrically double methylated arginine peptide | | Descriptor: | 1,2-ETHANEDIOL, DA2-ILE-PHE-SER peptide, E3 ubiquitin-protein ligase UBR2, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
6K8J
 
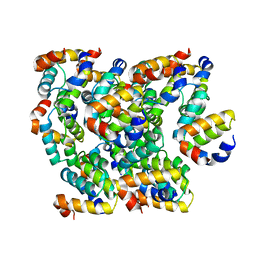 | | Structure of NLRC4 CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Gong, Q, Xu, C, Zhang, J, Boo, Z.Z, Wu, B. | | Deposit date: | 2019-06-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8.
Nat Commun, 12, 2021
|
|
4AB3
 
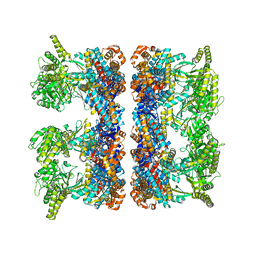 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
3D0J
 
 | | Crystal structure of conserved protein of unknown function CA_C3497 from Clostridium acetobutylicum ATCC 824 | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein CA_C3497 | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of Conserved Protein of Unknown Function CA_C3497 from Clostridium acetobutylicum ATCC 824.
To be Published
|
|
5TAT
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TBJ
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1452 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
