2CVS
 
 | | Structures of Yeast Ribonucleotide Reductase I | | 分子名称: | Ribonucleoside-diphosphate reductase large chain 1 | | 著者 | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | 登録日 | 2005-06-14 | | 公開日 | 2006-03-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1KZG
 
 | | DbsCdc42(Y889F) | | 分子名称: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | 著者 | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | 登録日 | 2002-02-06 | | 公開日 | 2002-03-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
3AQ5
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Fe(II)-O2 form | | 分子名称: | Group 1 truncated hemoglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Igarashi, J, Kobayashi, K, Matsuoka, A. | | 登録日 | 2010-10-25 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AR6
 
 | | Calcium pump crystal structure with bound TNP-ADP and TG in the absence of calcium | | 分子名称: | 2',3'-O-[(1R,6R)-2,4,6-trinitrocyclohexa-2,4-diene-1,1-diyl]adenosine 5'-(trihydrogen diphosphate), MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | 著者 | Toyoshima, C, Yonekura, S, Tsueda, J, Iwasawa, S. | | 登録日 | 2010-11-24 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1L4W
 
 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | 分子名称: | Acetylcholine receptor protein, alpha-Bungarotoxin | | 著者 | Samson, A.O, Scherf, T, Rodriguez, E, Eisenstein, M, Anglister, J. | | 登録日 | 2002-03-06 | | 公開日 | 2002-07-17 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1OX3
 
 | |
2CEU
 
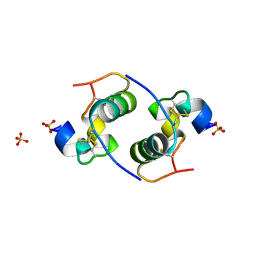 | | Despentapeptide insulin in acetic acid (pH 2) | | 分子名称: | INSULIN, SULFATE ION | | 著者 | Whittingham, J.L, Zhang, Y, Zakova, L, Dodson, E.J, Turkenburg, J.P, Brange, J, Dodson, G.G. | | 登録日 | 2006-02-10 | | 公開日 | 2006-03-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | I222 Crystal Form of Despentapeptide (B26-B30) Insulin Provides New Insights Into the Properties of Monomeric Insulin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2CCM
 
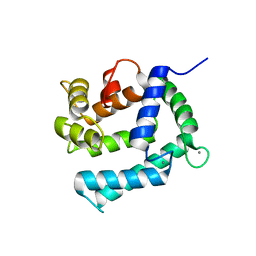 | | X-ray structure of Calexcitin from Loligo pealeii at 1.8A | | 分子名称: | CALCIUM ION, CALEXCITIN | | 著者 | Erskine, P.T, Beaven, G.D.E, Wood, S.P, Fox, G, Vernon, J, Giese, K.P, Cooper, J.B. | | 登録日 | 2006-01-16 | | 公開日 | 2006-01-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the Neuronal Protein Calexcitin Suggests a Mode of Interaction in Signalling Pathways of Learning and Memory.
J.Mol.Biol., 357, 2006
|
|
1LD6
 
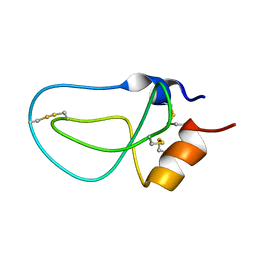 | | STRUCTURE OF BPTI_8A MUTANT | | 分子名称: | PANCREATIC TRYPSIN INHIBITOR | | 著者 | Cierpicki, T, Otlewski, J. | | 登録日 | 2002-04-08 | | 公開日 | 2002-09-11 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structures of two variants of bovine pancreatic trypsin inhibitor (BPTI) reveal unexpected influence of mutations on protein structure and stability.
J.Mol.Biol., 321, 2002
|
|
2CAZ
 
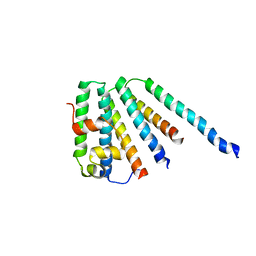 | | ESCRT-I core | | 分子名称: | PROTEIN SRN2, SUPPRESSOR PROTEIN STP22 OF TEMPERATURE-SENSITIVE ALPHA-FACTOR RECEPTOR AND ARGININE PERMEASE, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN VPS28 | | 著者 | Gill, D.J, Teo, H, Sun, J, Perisic, O, Veprintsev, D.B, Vallis, Y, Emr, S.D, Williams, R.L. | | 登録日 | 2005-12-23 | | 公開日 | 2006-04-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
1P9C
 
 | |
2CNJ
 
 | | NMR studies on the interaction of Insulin-Growth Factor II (IGF-II) with IGF2R domain 11 | | 分子名称: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | 著者 | Williams, C, Prince, S, Zaccheo, O, Hassan, A.B, Crosby, J, Crump, M. | | 登録日 | 2006-05-22 | | 公開日 | 2007-05-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights Into the Interaction of Insulin-Like Growth Factor 2 with Igf2R Domain 11.
Structure, 15, 2007
|
|
2JKV
 
 | | Structure of human Phosphogluconate Dehydrogenase in complex with NADPH at 2.53A | | 分子名称: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING, CHLORIDE ION, ... | | 著者 | Pilka, E.S, Kavanagh, K.L, von Delft, F, Muniz, J.R.C, Murray, J, Picaud, S, Guo, K, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Oppermann, U. | | 登録日 | 2008-09-01 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.532 Å) | | 主引用文献 | Structure of Human Phosphogluconate Dehydrogenase in Complex with Nadph at 2.53A
To be Published
|
|
2D49
 
 | | Solution structure of the Chitin-Binding Domain of Streptomyces griseus Chitinase C | | 分子名称: | chitinase C | | 著者 | Akagi, K, Watanabe, J, Hara, M, Kezuka, Y, Chikaishi, E, Yamaguchi, T, Akutsu, H, Nonaka, T, Watanabe, T, Ikegami, T. | | 登録日 | 2005-10-11 | | 公開日 | 2006-10-11 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C
J.Biochem.(Tokyo), 139, 2006
|
|
2D1Q
 
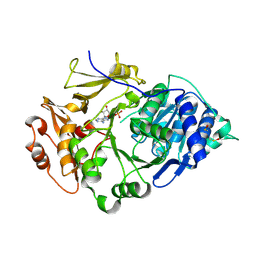 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with MgATP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | 著者 | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-08-31 | | 公開日 | 2006-03-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D6B
 
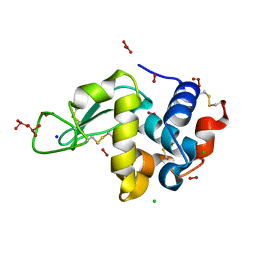 | |
1Q2B
 
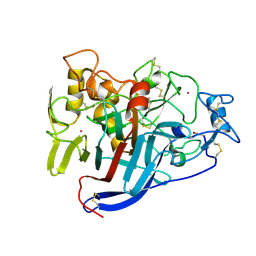 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | 著者 | Stahlberg, J, Harris, M, Jones, T.A. | | 登録日 | 2003-07-24 | | 公開日 | 2003-11-25 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
5V2X
 
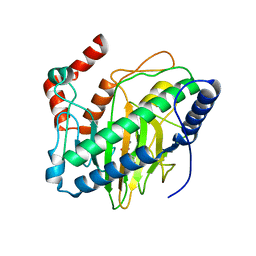 | | Ethylene forming enzyme in complex with manganese and 2-oxoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, MANGANESE (II) ION | | 著者 | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | 登録日 | 2017-03-06 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.847 Å) | | 主引用文献 | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
2D1R
 
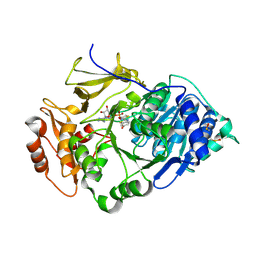 | | Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP | | 分子名称: | 2-(6-HYDROXY-1,3-BENZOTHIAZOL-2-YL)-1,3-THIAZOL-4(5H)-ONE, ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | 著者 | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-08-31 | | 公開日 | 2006-03-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D26
 
 | |
2D41
 
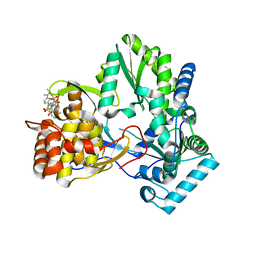 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor | | 分子名称: | 5'-ACETYL-4-{[(2,4-DIMETHYLPHENYL)SULFONYL]AMINO}-2,2'-BITHIOPHENE-5-CARBOXYLIC ACID, polyprotein | | 著者 | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | 登録日 | 2005-10-05 | | 公開日 | 2006-08-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
1Q5W
 
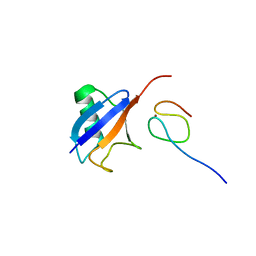 | | Ubiquitin Recognition by Npl4 Zinc-Fingers | | 分子名称: | Ubiquitin, ZINC ION, homolog of yeast nuclear protein localization 4 | | 著者 | Alam, S.L, Sun, J, Payne, M, Welch, B.D, Blake, B.K, Davis, D.R, Meyer, H.H, Emr, S.D, Sundquist, W.I. | | 登録日 | 2003-08-11 | | 公開日 | 2004-03-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Ubiquitin interactions of NZF zinc fingers.
Embo J., 23, 2004
|
|
5UK1
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
3B5A
 
 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating A38G mutation with a 2'OMe modification at the active site | | 分子名称: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | 著者 | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | 登録日 | 2007-10-25 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B7O
 
 | | Crystal structure of the human tyrosine phosphatase SHP2 (PTPN11) with an accessible active site | | 分子名称: | D-MALATE, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Ugochukwu, E, Barr, A, Patel, A, King, O, Niesen, F, Salah, E, Savitsky, P, Pilka, E.S, Elkins, J, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2007-10-31 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
