4ZEA
 
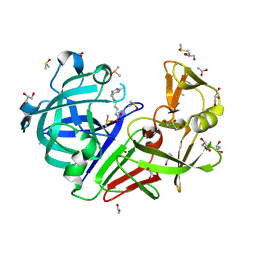 | | Endothiapepsin in complex with fragment B91 | | 分子名称: | 1,2-ETHANEDIOL, 2-IMINOBIOTIN, ACETATE ION, ... | | 著者 | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | 登録日 | 2015-04-20 | | 公開日 | 2016-05-04 | | 最終更新日 | 2016-05-11 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
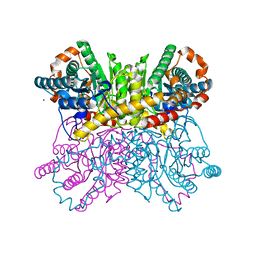
4ZB0
 
 | |
4O9K
 
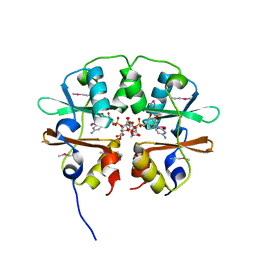 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | 分子名称: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | 著者 | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-01-02 | | 公開日 | 2014-01-22 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
4O9C
 
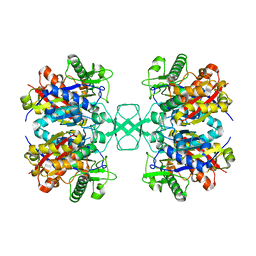 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | 分子名称: | Acetyl-CoA acetyltransferase, COENZYME A | | 著者 | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | 登録日 | 2014-01-02 | | 公開日 | 2014-12-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4G6H
 
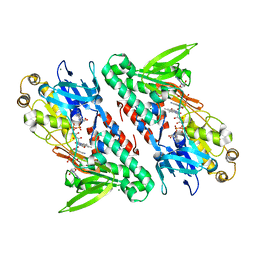 | | Crystal structure of NDH with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | 著者 | Li, W, Feng, Y, Ge, J, Yang, M. | | 登録日 | 2012-07-19 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.262 Å) | | 主引用文献 | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
6QJY
 
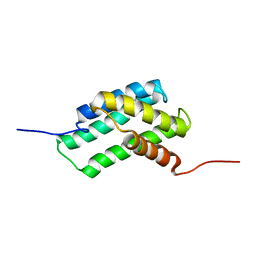 | |
5L18
 
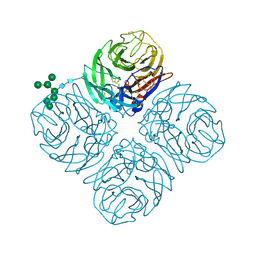 | |
7JLT
 
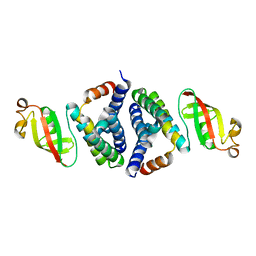 | |
3HPM
 
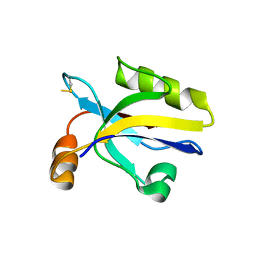 | |
4ZN9
 
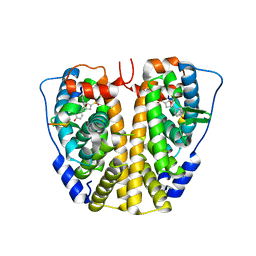 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Oxabicyclic Heptene Sulfonate (OBHS) | | 分子名称: | Estrogen receptor, Nuclear receptor-interacting peptide, cyclohexa-2,5-dien-1-yl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-05-04 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.215 Å) | | 主引用文献 | Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands.
Chemmedchem, 7, 2012
|
|
4CY2
 
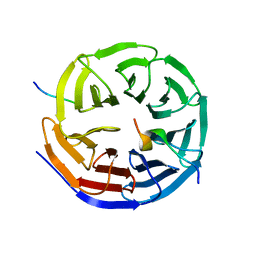 | | Crystal structure of the KANSL1-WDR5-KANSL2 complex. | | 分子名称: | KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, KAT8 REGULATORY NSL COMPLEX SUBUNIT 2, WD REPEAT-CONTAINING PROTEIN 5 | | 著者 | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | 登録日 | 2014-04-09 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
6I7D
 
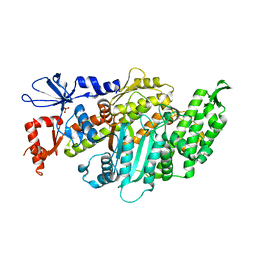 | | Plasmodium falciparum Myosin A, post-rigor and rigor-like states | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Myosin-A | | 著者 | Robert-Paganin, J, Auguin, D, Moussaoui, D, Jousset, G, Baum, J, Trybus, K.M, Houdusse, A. | | 登録日 | 2018-11-16 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism.
Nat Commun, 10, 2019
|
|
2RCY
 
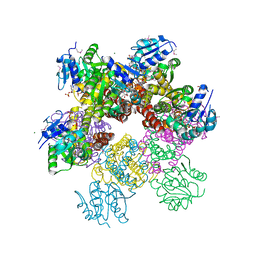 | | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound | | 分子名称: | GLYCEROL, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Wernimont, A.K, Lew, J, Lin, Y.H, Ren, H, Sun, X, Khuu, C, Hassanali, A, Wasney, G, Zhao, Y, Kozieradzki, I, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2007-09-20 | | 公開日 | 2007-10-23 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound.
To be Published
|
|
4OQ3
 
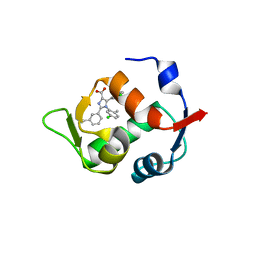 | |
8EPG
 
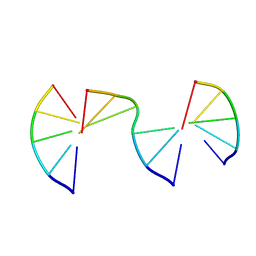 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | 分子名称: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*C)-3') | | 著者 | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | 登録日 | 2022-10-05 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8F42
 
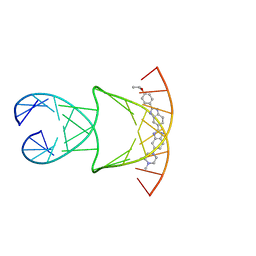 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | 分子名称: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*TP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*GP*C)-3') | | 著者 | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPI
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | 分子名称: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*TP*A)-3') | | 著者 | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | 登録日 | 2022-10-05 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPE
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | 分子名称: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*G)-3') | | 著者 | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | 登録日 | 2022-10-05 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPB
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | 分子名称: | DNA (5'-D(*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*A)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*T)-3') | | 著者 | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | 登録日 | 2022-10-05 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
6I54
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 2. | | 分子名称: | Influenza virus nucleoprotein, Nucleoprotein | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-11-12 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
4ZOW
 
 | | Crystal structure of E. coli multidrug transporter MdfA in complex with chloramphenicol | | 分子名称: | CHLORAMPHENICOL, Multidrug transporter MdfA | | 著者 | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | 登録日 | 2015-05-07 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
8F40
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | 分子名称: | DNA (5'-D(*CP*GP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*GP*C)-3') | | 著者 | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
5DP3
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 2 | | 分子名称: | 3C proteinase, ethyl (4S)-5-[(3S)-2-oxopyrrolidin-3-yl]-4-[(3-phenylpropanoyl)amino]pent-2-enoate | | 著者 | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | 登録日 | 2015-09-12 | | 公開日 | 2016-03-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
3I0M
 
 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | 分子名称: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | 著者 | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | 登録日 | 2009-06-25 | | 公開日 | 2009-10-13 | | 最終更新日 | 2012-05-02 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
