1RVI
 
 | | SOLUTION STRUCTURE OF THE DNA DODECAMER CGTTTTAAAACG | | 分子名称: | 5'-D(*CP*GP*TP*TP*TP*TP*AP*AP*AP*AP*CP*G)-3' | | 著者 | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | 登録日 | 2003-12-13 | | 公開日 | 2004-02-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4A9T
 
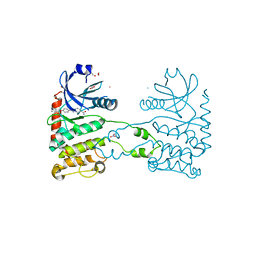 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | 分子名称: | 1,2-ETHANEDIOL, 2-(4-{[1-(4-CHLOROBENZYL)PIPERIDIN-4-YL]METHOXY}PHENYL)-1H-BENZIMIDAZOLE-5-CARBOXAMIDE, NITRATE ION, ... | | 著者 | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | 登録日 | 2011-11-28 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography.
Bioorg.Med.Chem., 20, 2012
|
|
4AAO
 
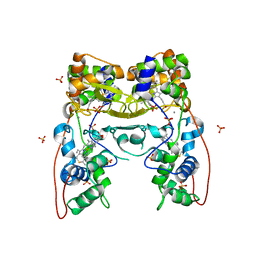 | | MacA-H93G | | 分子名称: | CALCIUM ION, CYTOCHROME C551 PEROXIDASE, HEME C, ... | | 著者 | Seidel, J. | | 登録日 | 2011-12-05 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Maca is a Second Cytochrome C Peroxidase of Geobacter Sulfurreducens.
Biochemistry, 51, 2012
|
|
1RS7
 
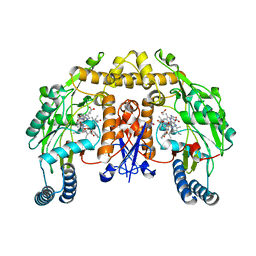 | | Rat neuronal NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | 著者 | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | 登録日 | 2003-12-09 | | 公開日 | 2004-05-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
3HHG
 
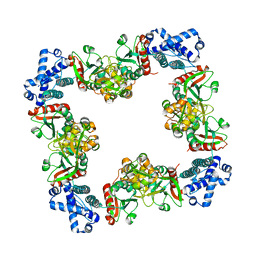 | | Structure of CrgA, a LysR-type transcriptional regulator from Neisseria meningitidis. | | 分子名称: | Transcriptional regulator, LysR family | | 著者 | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | 登録日 | 2009-05-15 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
1Q0R
 
 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin T (DcmaT) | | 分子名称: | 10-DECARBOXYMETHYLACLACINOMYCIN T (DCMAT), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | 著者 | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | 登録日 | 2003-07-17 | | 公開日 | 2003-11-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
3HKO
 
 | | Crystal structure of a cdpk kinase domain from cryptosporidium Parvum, cgd7_40 | | 分子名称: | Calcium/calmodulin-dependent protein kinase with a kinase domain and 2 calmodulin-like EF hands, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Wernimont, A.K, Hutchinson, A, Wasney, G, Vedadi, M, MacKenzie, F, Kozieradzki, I, Cossar, D, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Botchkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2009-05-25 | | 公開日 | 2009-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a cdpk kinase domain from cryptosporidium Parvum, cgd7_40
To be Published
|
|
4ALI
 
 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | 分子名称: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ACZ
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron | | 分子名称: | ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | 著者 | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | 登録日 | 2011-12-21 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1ROT
 
 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | FKBP59-I | | 著者 | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | 登録日 | 1996-06-14 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
3IYX
 
 | |
1ROU
 
 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, 22 STRUCTURES | | 分子名称: | FKBP59-I | | 著者 | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | 登録日 | 1996-06-14 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
3UPL
 
 | | Crystal structure of the Brucella abortus enzyme catalyzing the first committed step of the methylerythritol 4-phosphate pathway. | | 分子名称: | GLYCEROL, MAGNESIUM ION, Oxidoreductase | | 著者 | Calisto, B.M, Perez-Gil, J, Fita, I, Rodriguez-Concepcion, M. | | 登録日 | 2011-11-18 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of Brucella abortus deoxyxylulose-5-phosphate reductoisomerase-like (DRL) enzyme involved in isoprenoid biosynthesis.
J.Biol.Chem., 287, 2012
|
|
3IST
 
 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid | | 分子名称: | CHLORIDE ION, Glutamate racemase, SUCCINIC ACID | | 著者 | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-08-27 | | 公開日 | 2009-09-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid
TO BE PUBLISHED
|
|
3IT7
 
 | | Crystal Structure of the LasA virulence factor from Pseudomonas aeruginosa | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, Protease lasA, ... | | 著者 | Spencer, J, Murphy, L.M, Conners, R, Sessions, R.B, Gamblin, S.J. | | 登録日 | 2009-08-27 | | 公開日 | 2009-11-17 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Crystal structure of the LasA virulence factor from Pseudomonas aeruginosa: substrate specificity and mechanism of M23 metallopeptidases.
J.Mol.Biol., 396, 2010
|
|
3V72
 
 | |
3USI
 
 | |
3V7B
 
 | | Dip2269 protein from corynebacterium diphtheriae | | 分子名称: | 1,2-ETHANEDIOL, Uncharacterized protein | | 著者 | Osipiuk, J, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-12-20 | | 公開日 | 2012-01-11 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.743 Å) | | 主引用文献 | Dip2269 protein from corynebacterium diphtheriae.
To be Published
|
|
3IV1
 
 | | Coiled-coil domain of tumor susceptibility gene 101 | | 分子名称: | CHLORIDE ION, SULFATE ION, Tumor susceptibility gene 101 protein | | 著者 | Neculai, D, Avvakumov, G.V, Wernimont, A.K, Xue, S, Walker, J.R, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2009-08-31 | | 公開日 | 2009-11-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Coiled-Coil Domain of Human Tsg101
To be Published
|
|
3ULV
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | 著者 | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | 登録日 | 2011-11-11 | | 公開日 | 2012-05-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.522 Å) | | 主引用文献 | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3UWA
 
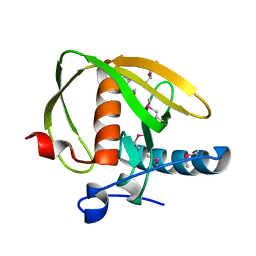 | | Crystal structure of a probable peptide deformylase from synechococcus phage S-SSM7 | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, RIIA-RIIB membrane-associated protein, ZINC ION | | 著者 | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | 登録日 | 2011-12-01 | | 公開日 | 2013-01-09 | | 最終更新日 | 2013-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
1RSU
 
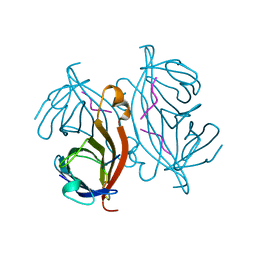 | |
3UOW
 
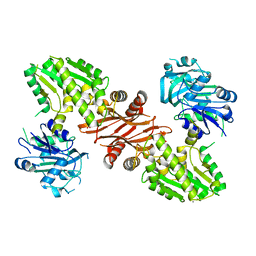 | | Crystal Structure of PF10_0123, a GMP Synthetase from Plasmodium Falciparum | | 分子名称: | CALCIUM ION, GMP synthetase, XANTHOSINE-5'-MONOPHOSPHATE | | 著者 | Wernimont, A.K, Dong, A, Hills, T, Amani, M, Perieteanu, A, Lin, Y.H, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Structural Genomics Consortium (SGC) | | 登録日 | 2011-11-17 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Crystal Structure of PF10_0123, a GMP Synthetase from Plasmodium Falciparum
TO BE PUBLISHED
|
|
1RNB
 
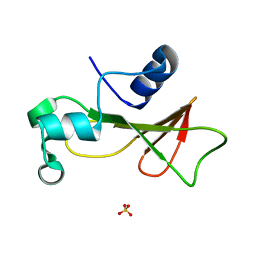 | |
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | 著者 | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | 登録日 | 2009-08-01 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (19.799999 Å) | | 主引用文献 | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
