5XQW
 
 | | Catalytic antibody 7B9 | | Descriptor: | Fab fragment of catalytic antibody 7B9, heavy chain, light chain, ... | | Authors: | Ito, N, Fujii, I, Tsumuraya, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the broad substrate tolerance of the antibody 7B9-catalyzed hydrolysis of p-nitrobenzyl esters.
Bioorg. Med. Chem., 26, 2018
|
|
1V33
 
 | | Crystal structure of DNA primase from Pyrococcus horikoshii | | Descriptor: | DNA primase small subunit, PHOSPHATE ION, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
1V34
 
 | | Crystal structure of Pyrococcus horikoshii DNA primase-UTP complex | | Descriptor: | DNA primase small subunit, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
2Z8H
 
 | | Structure of mouse Bach1 BTB domain | | Descriptor: | Transcription regulator protein BACH1 | | Authors: | Ito, N, Murayama, K. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of mouse Bach1 BTB domain
To be Published
|
|
1GOH
 
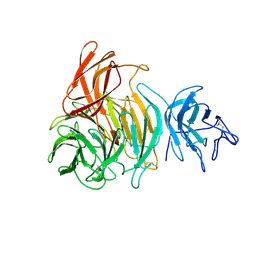 | |
1GOG
 
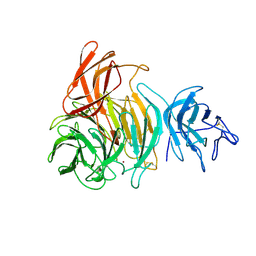 | |
1GOF
 
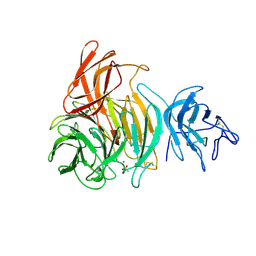 | | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE | | Descriptor: | ACETIC ACID, COPPER (II) ION, GALACTOSE OXIDASE, ... | | Authors: | Ito, N, Phillips, S.E.V, Knowles, P.F. | | Deposit date: | 1993-09-30 | | Release date: | 1994-01-31 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase.
Nature, 350, 1991
|
|
2DLA
 
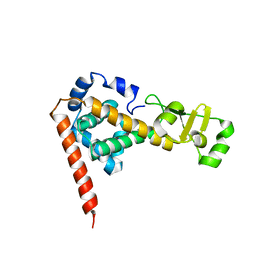 | |
1HBH
 
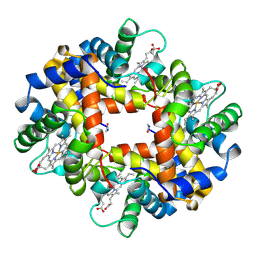 | |
7E0D
 
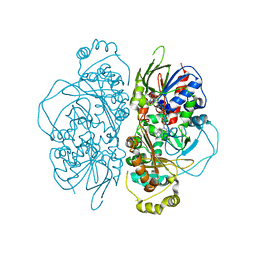 | | Structure of L-glutamate oxidase R305E mutant in complex with L-arginine | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
7E0C
 
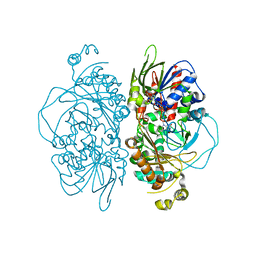 | | Structure of L-glutamate oxidase R305E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
7D4C
 
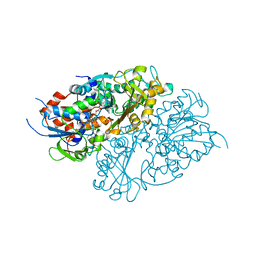 | | Structure of L-lysine oxidase precursor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, PHOSPHATE ION | | Authors: | Ito, N, Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
3VYC
 
 | |
8YTN
 
 | | Single-chain Fv antibody of E11 | | Descriptor: | GLYCEROL, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTO
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTP
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine under reducing conditions | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
6LCB
 
 | | Crystal structure of human Dishevelled1 PDZ domain with its inhibitor NPL3009 | | Descriptor: | 2-[[3-[(2E)-2-[1,3-bis(oxidanylidene)-1-phenyl-butan-2-ylidene]hydrazinyl]phenyl]sulfonylamino]benzoic acid, SULFATE ION, Segment polarity protein dishevelled homolog DVL-1 | | Authors: | Yasukochi, S, Numoto, N, Tenno, N, Tenno, T, Ito, N, Hiroaki, H. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of human Dishevelled1 PDZ domain with its inhibitor NPL3009
To Be Published
|
|
6K4Z
 
 | | Single-chain Fv antibody of C6 COMPLEXED WITH 2-(4-HYDROXY-3-NITROPHENYL)ACETIC ACID | | Descriptor: | 2-(4-HYDROXY-3-NITROPHENYL)ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Nishiguchi, A, Numoto, N, Ito, N, Azuma, T, Oda, M. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional structure of a high affinity anti-(4-hydroxy-3-nitrophenyl)acetyl antibody possessing a glycine residue at position 95 of the heavy chain.
Mol.Immunol., 114, 2019
|
|
6LB5
 
 | | Crystal structure of dimeric RXR-LBD complexed with full agonist NEt-3IB and TIF2 co-activator | | Descriptor: | 6-[ethyl-[3-(2-methylpropoxy)-4-propan-2-yl-phenyl]amino]pyridine-3-carboxylic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Imai, D, Numoto, N, Nakano, S, Kakuta, H, Ito, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of dimeric RXR-LBD complexed with full agonist NEt-3IB and TIF2 co-activator
To Be Published
|
|
7CTR
 
 | | Closed form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
7C7W
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3S,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
7CTS
 
 | | Open form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein, BICINE, ... | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
6K5O
 
 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Kagechika, H, Ito, N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
7CIO
 
 | | Molecular interactions of cytoplasmic region of CTLA-4 with SH2 domains of PI3-kinase | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Iiyama, M, Numoto, N, Ogawa, S, Kuroda, M, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular interactions of the CTLA-4 cytoplasmic region with the phosphoinositide 3-kinase SH2 domains.
Mol.Immunol., 131, 2021
|
|
7CEF
 
 | | Crystal structure of PET-degrading cutinase Cut190 /S226P/R228S/ mutant with the C-terminal three residues deletion | | Descriptor: | Alpha/beta hydrolase family protein, CALCIUM ION, ZINC ION | | Authors: | Senga, A, Numoto, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-06-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
J.Biochem., 169, 2021
|
|
