8HAP
 
 | | Crystal structure of thermostable acetaldehyde dehydrogenase from hyperthermophilic archaeon Sulfolobus tokodaii | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, SODIUM ION, ... | | Authors: | Mine, S, Nakabayashi, M, Ishikawa, K. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable acetaldehyde dehydrogenase from the hyperthermophilic archaeon Sulfolobus tokodaii.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
5D48
 
 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | Descriptor: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | Descriptor: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5HXV
 
 | |
4DM1
 
 | |
4DM2
 
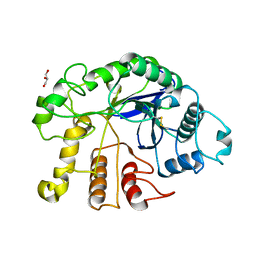 | |
7X8V
 
 | | Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity | | Descriptor: | Os01g0156300 protein | | Authors: | Ichimaru, K, Harada, K, Yamaguchi, K, Shigeta, S, Shimada, K, Ishikawa, K, Inoue, K, Nishio, Y, Yoshimura, S, Inoue, H, Yamashita, E, Fujiwara, T, Nakagawa, A, Kojima, C, Kawasaki, T. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity.
Nat Commun, 13, 2022
|
|
1WKV
 
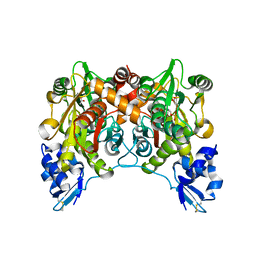 | | Crystal structure of O-phosphoserine sulfhydrylase | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, cysteine synthase | | Authors: | Oda, Y, Mino, K, Ishikawa, K, Ataka, M. | | Deposit date: | 2004-06-09 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional Structure of a New Enzyme, O-Phosphoserine Sulfhydrylase, involved in l-Cysteine Biosynthesis by a Hyperthermophilic Archaeon, Aeropyrum pernix K1, at 2.0A Resolution
J.Mol.Biol., 351, 2005
|
|
1X0R
 
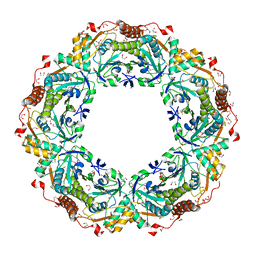 | | Thioredoxin Peroxidase from Aeropyrum pernix K1 | | Descriptor: | 1,2-ETHANEDIOL, Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Inoue, T, Matsumura, H, Kobayashi, A, Hagihara, Y, Uegaki, K, Ataka, M, Kai, Y, Ishikawa, K. | | Deposit date: | 2005-03-28 | | Release date: | 2005-12-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of thioredoxin peroxidase from aerobic hyperthermophilic archaeon Aeropyrum pernix K1
Proteins, 62, 2006
|
|
5B5S
 
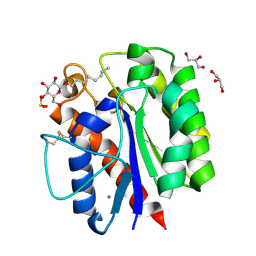 | |
2DSK
 
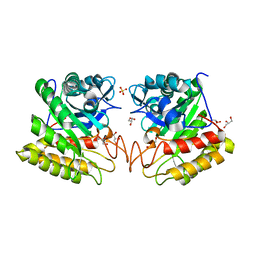 | | Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, chitinase | | Authors: | Nakamura, T, Mine, S, Hagihara, Y, Ishikawa, K, Uegaki, K. | | Deposit date: | 2006-06-30 | | Release date: | 2007-02-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the catalytic domain of the hyperthermophilic chitinase from Pyrococcus furiosus
ACTA CRYSTALLOGR.,SECT.F, 63, 2007
|
|
1IU4
 
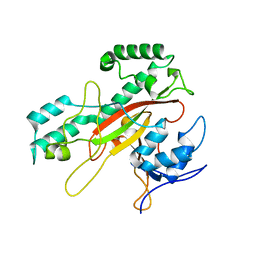 | | Crystal Structure Analysis of the Microbial Transglutaminase | | Descriptor: | microbial transglutaminase | | Authors: | Kashiwagi, T, Yokoyama, K, Ishikawa, K, Ono, K, Ejima, D, Matsui, H, Suzuki, E. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense
J.Biol.Chem., 277, 2002
|
|
2CWR
 
 | |
2CZN
 
 | | Solution structure of the chitin-binding domain of hyperthermophilic chitinase from pyrococcus furiosus | | Descriptor: | chitinase | | Authors: | Uegaki, T, Ikegami, T, Nakamura, T, Hagihara, Y, Mine, S, Inoue, T, Matsumura, H, Ataka, M, Ishikawa, K. | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Mol.Biol., 381, 2008
|
|
1WMU
 
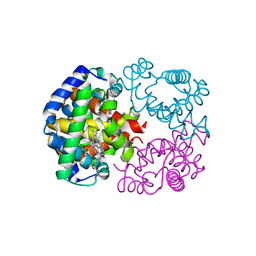 | | Crystal Structure of Hemoglobin D from the Aldabra Giant Tortoise, Geochelone gigantea, at 1.65 A resolution | | Descriptor: | Hemoglobin A and D beta chain, Hemoglobin D alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Satoh, I, Ishikawa, K, Shishikura, F. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Hemoglobin D from the Aldabra Giant Tortoise, Geochelone gigantea, at 1.65 A resolution
To be Published
|
|
1X3K
 
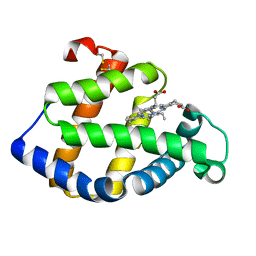 | | Crystal structure of a hemoglobin component (TA-V) from Tokunagayusurika akamusi | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin component V | | Authors: | Kuwada, T, Hasegawa, T, Sato, S, Sato, I, Ishikawa, K, Takagi, T, Shishikura, F. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of two hemoglobin components from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera).
Gene, 398, 2007
|
|
1X46
 
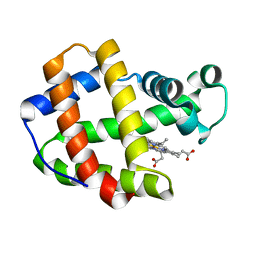 | | Crystal structure of a hemoglobin component (TA-VII) from Tokunagayusurika akamusi | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin component VII | | Authors: | Kuwada, T, Hasegawa, T, Sato, S, Sato, I, Ishikawa, K, Takagi, T, Shishikura, F. | | Deposit date: | 2005-05-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of two hemoglobin components from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera).
Gene, 398, 2007
|
|
7V92
 
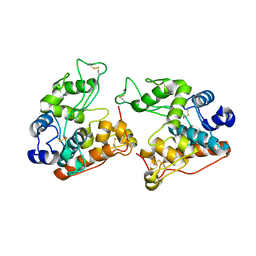 | |
7V91
 
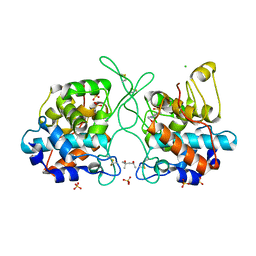 | | Crystal Structure of the Catalytic Domain of a Family GH19 Chitinase from Gazyumaru, Ficus microcarpa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kozome, D, Kubota, T, Ishikawa, K. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis and Construction of a Thermostable Antifungal Chitinase.
Appl.Environ.Microbiol., 88, 2022
|
|
1V75
 
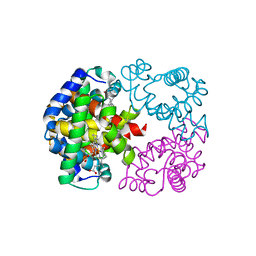 | | Crystal structure of hemoglobin D from the Aldabra giant tortoise (Geochelone gigantea) at 2.0 A resolution | | Descriptor: | Hemoglobin A and D beta chain, Hemoglobin D alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Satoh, I, Ishikawa, K, Shishikura, F. | | Deposit date: | 2003-12-12 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallization and preliminary X-ray diffraction study of hemoglobin D from the Aldabra giant tortoise, Geochelone gigantea.
Protein Pept.Lett., 10, 2003
|
|
3VSA
 
 | | Crystal Structure of O-phosphoserine sulfhydrylase without acetate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PYRIDOXAL-5'-PHOSPHATE, Protein CysO | | Authors: | Nakamura, T, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural analysis of the substrate recognition mechanism in O-phosphoserine sulfhydrylase from the hyperthermophilic archaeon Aeropyrum pernix K1
J.Mol.Biol., 422, 2012
|
|
3A4X
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
3A4W
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, MAGNESIUM ION, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
