8IHF
 
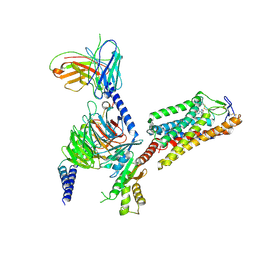 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | 分子名称: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
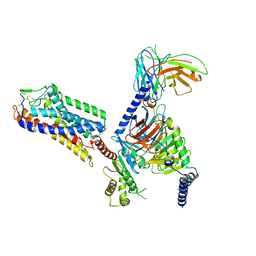 | | Cryo-EM structure of HCA3-Gi complex with acifran | | 分子名称: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHB
 
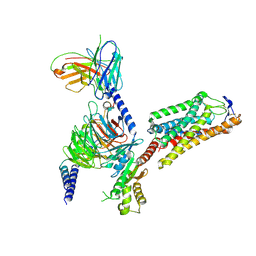 | | Cryo-EM structure of HCA2-Gi complex with GSK256073 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
3X29
 
 | | CRYSTAL STRUCTURE of MOUSE CLAUDIN-19 IN COMPLEX with C-TERMINAL FRAGMENT OF CLOSTRIDIUM PERFRINGENS ENTEROTOXIN | | 分子名称: | Claudin-19, Heat-labile enterotoxin B chain | | 著者 | Saitoh, Y, Suzuki, H, Tani, K, Nishikawa, K, Irie, K, Ogura, Y, Tamura, A, Tsukita, S, Fujiyoshi, Y. | | 登録日 | 2014-12-13 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin
Science, 347, 2015
|
|
2Z1H
 
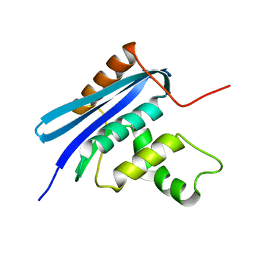 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | 分子名称: | Ribonuclease HI | | 著者 | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-05-08 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1G
 
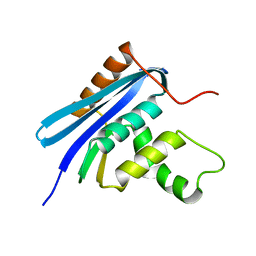 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K) | | 分子名称: | Ribonuclease HI | | 著者 | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-05-08 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2ZZ9
 
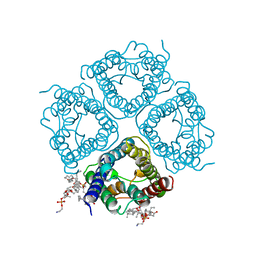 | | Structure of aquaporin-4 S180D mutant at 2.8 A resolution by electron crystallography | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aquaporin-4 | | 著者 | Tani, K, Mitsuma, T, Hiroaki, Y, Kamegawa, A, Nishikawa, K, Tanimura, Y, Fujiyoshi, Y. | | 登録日 | 2009-02-06 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | 主引用文献 | Mechanism of Aquaporin-4's Fast and Highly Selective Water Conduction and Proton Exclusion.
J.Mol.Biol., 389, 2009
|
|
2Z1I
 
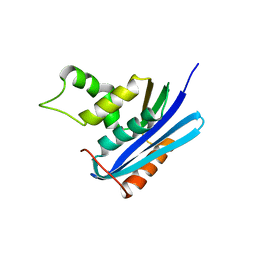 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K) | | 分子名称: | Ribonuclease HI | | 著者 | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-05-08 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1J
 
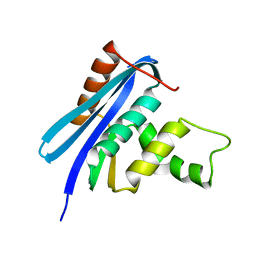 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | 分子名称: | Ribonuclease HI | | 著者 | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | 登録日 | 2007-05-08 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
