8V5T
 
 | | Crystal structure of Alzheimers disease phospholipase D3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, GLYCEROL, ... | | 著者 | Ishii, K, Hermans, S.J, Nero, T.L, Gorman, M.A, Parker, M.W. | | 登録日 | 2023-12-01 | | 公開日 | 2024-10-09 | | 最終更新日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of Alzheimer's disease phospholipase D3 provides a molecular basis for understanding its normal and pathological functions.
Febs J., 291, 2024
|
|
5ZRQ
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in Zn(2+)-bound state | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZNO
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant in Ca(2+)-bound state | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL | | 著者 | Numoto, N, Inaba, S, Yamagami, Y, Kamiya, N, Bekker, G.J, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-10 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.60264349 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZRS
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl adipate bound state | | 分子名称: | 6-ethoxy-6-oxohexanoic acid, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZRR
 
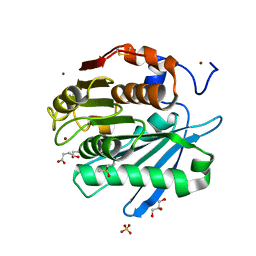 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl succinate bound state | | 分子名称: | 4-ethoxy-4-oxobutanoic acid, Alpha/beta hydrolase family protein, GLYCEROL, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
3ASP
 
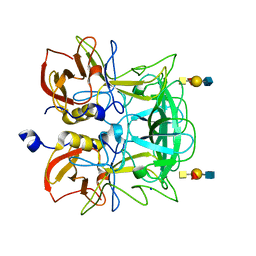 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with A-antigen | | 分子名称: | Capsid protein, SODIUM ION, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | 登録日 | 2010-12-17 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3AST
 
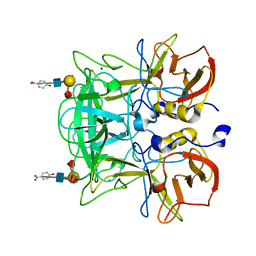 | | Crystal structure of P domain Q389N mutant from Norovirus Funabashi258 stain in the complex with Lewis-b | | 分子名称: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | 著者 | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | 登録日 | 2010-12-17 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASQ
 
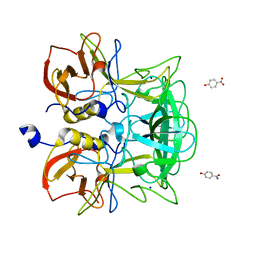 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with H-antigen | | 分子名称: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | 著者 | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | 登録日 | 2010-12-17 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASR
 
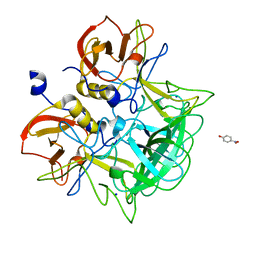 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-a | | 分子名称: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | 著者 | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | 登録日 | 2010-12-17 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASS
 
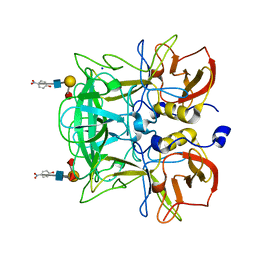 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-b | | 分子名称: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | 著者 | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | 登録日 | 2010-12-17 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ALT
 
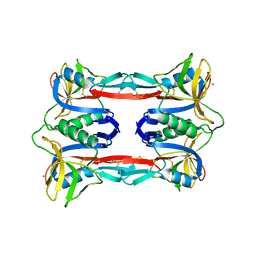 | | Crystal structure of CEL-IV complexed with Melibiose | | 分子名称: | CALCIUM ION, Lectin CEL-IV, C-type, ... | | 著者 | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | 登録日 | 2010-08-07 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALU
 
 | | Crystal structure of CEL-IV complexed with Raffinose | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Lectin CEL-IV, ... | | 著者 | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | 登録日 | 2010-08-07 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALS
 
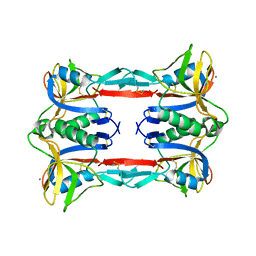 | | Crystal structure of CEL-IV | | 分子名称: | CALCIUM ION, Lectin CEL-IV, C-type | | 著者 | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | 登録日 | 2010-08-07 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
