4DZI
 
 | | Crystal structure of amidohydrolase map2389c (target EFI-500390) from Mycobacterium avium subsp. paratuberculosis K-10 | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of amidohydrolase map2389c (target EFI-500390) from Mycobacterium avium
To be Published
|
|
4IW9
 
 | | Crystal structure of glutathione s-transferase mha_0454 (target efi-507015) from mannheimia haemolytica, gsh complex | | Descriptor: | ACETATE ION, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Probable Glutathione S-Transferase Mha_0454 (Target Efi-507015) from Mannheimia Haemolytica
To be Published
|
|
4IQ1
 
 | | Crystal structure of glutathione s-transferase MHA_0454 (TARGET EFI-507015) FROM Mannheimia haemolytica, SUBSTRATE-FREE | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, AL Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of probable glutathione s-transferase MHA_0454 (TARGET EFI-507015) FROM Mannheimia haemolytica, Substrate-free
To be Published
|
|
4DHG
 
 | | Crystal structure of enolase TBIS_1083(TARGET EFI-502310) from Thermobispora bispora dsm 43833, an open loop conformation | | Descriptor: | GLYCEROL, IODIDE ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Enolase Tbis_1083 from Thermobispora Bispora Dsm 43833
To be Published
|
|
4JYX
 
 | | Crystal structure of polyprenyl synthase PATL_3739 (TARGET EFI-509195) FROM PSEUDOALTEROMONAS ATLANTICA, COMPLEX WITH INORGANIC PHOSPHATE AND AN UNKNOWN LIGAND | | Descriptor: | PHOSPHATE ION, Trans-hexaprenyltranstransferase, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-01 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase Patl_3739 from Pseudoalteromonas Atlantica
To be Published
|
|
4DN1
 
 | | Crystal structure of an ENOLASE (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate | | Descriptor: | CHLORIDE ION, FORMIC ACID, Isomerase/lactonizing enzyme, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Bouvier, J.T, Wasserman, S.R, Morisco, L.L, Sojitra, S, Al Obaidi, N.F, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate
to be published
|
|
4E38
 
 | | Crystal structure of probable keto-hydroxyglutarate-aldolase from Vibrionales bacterium SWAT-3 (Target EFI-502156) | | Descriptor: | CHLORIDE ION, Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of probable keto-hydroxyglutarate-aldolase from Vibrionales bacterium SWAT-3 (Target EFI-502156)
To be Published
|
|
4DW8
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I | | Descriptor: | Haloacid dehalogenase-like hydrolase, SODIUM ION, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I
To be Published
|
|
4JBD
 
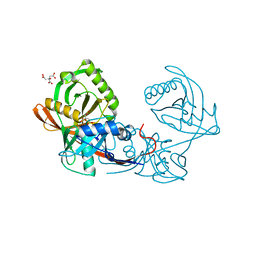 | | Crystal structure of Pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group I2, bound citrate | | Descriptor: | CITRIC ACID, Proline racemase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from pseudomonas putida f1 (target efi-506500), open form, space group i2, bound citrate
To be Published
|
|
4FI4
 
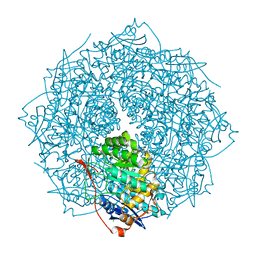 | | Crystal structure of mannonate dehydratase PRK15072 (TARGET EFI-502214) from Caulobacter sp. K31 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enolase Prk15072 (Target Efi-502214) from Caulobacter Sp. K31
To be Published
|
|
4E69
 
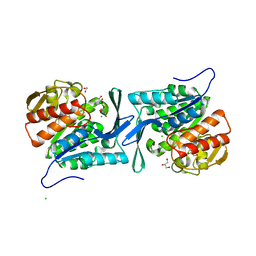 | | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydro-3-deoxygluconokinase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure
To be Published
|
|
4JUU
 
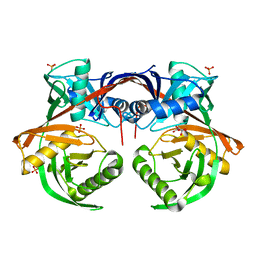 | | Crystal structure of a putative hydroxyproline epimerase from xanthomonas campestris (TARGET EFI-506516) with bound phosphate and unknown ligand | | Descriptor: | CHLORIDE ION, Epimerase, PHOSPHATE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative hydroxyproline epimerase from xanthomonas campestris (TARGET EFI-506516) with bound phosphate and unknown ligand
To be Published
|
|
4JXK
 
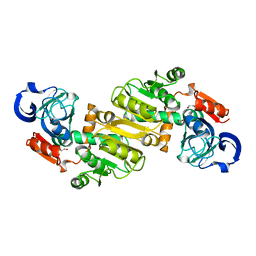 | | Crystal Structure of Oxidoreductase ROP_24000 (Target EFI-506400) from Rhodococcus opacus B4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, oxidoreductase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Oxidoreductase Rop_24000 (Target EFI-506400) from Rhodococcus Opacus
To be Published
|
|
4JXY
 
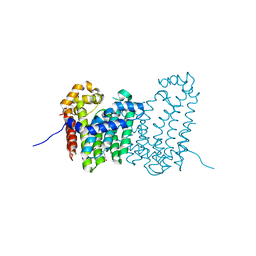 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE PATL_3739 (TARGET EFI-509195) FROM Pseudoalteromonas atlantica | | Descriptor: | GLUTARIC ACID, PHOSPHATE ION, Trans-hexaprenyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF ISOPRENOID SYNTHASE PATL_3739 FROM Pseudoalteromonas atlantica
To be Published
|
|
4JXN
 
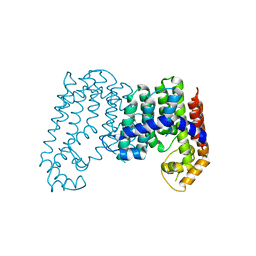 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE ISP_B (TARGET EFI-509198) FROM Roseobacter denitrificans | | Descriptor: | CHLORIDE ION, Octaprenyl-diphosphate synthase, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase Isp_B from Roseobacter Denitrificans
To be Published
|
|
4EEL
 
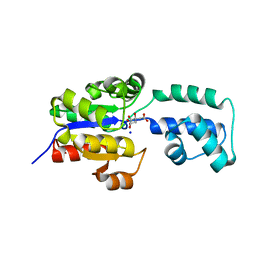 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound citrate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, CITRIC ACID, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4EEN
 
 | | crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound magnesium | | Descriptor: | Beta-phosphoglucomutase-related protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4EBU
 
 | | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-24 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I
TO BE PUBLISHED
|
|
4ECI
 
 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with acetate | | Descriptor: | ACETATE ION, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
4L8E
 
 | | Crystal structure of a glutathione transferase family member from xenorhabdus nematophila, target efi-507418, with two gsh per subunit | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-17 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a glutathione transferase family member from xenorhabdus nematophila, target efi-507418, with two gsh per subunit
To be published
|
|
4LB0
 
 | | Crystal structure of a hydroxyproline epimerase from agrobacterium vitis, target efi-506420, with bound trans-4-oh-l-proline | | Descriptor: | 4-HYDROXYPROLINE, ACETATE ION, Uncharacterized protein | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a hydroxyproline epimerase from agrobacterium vitis, target efi-506420, with bound trans-4-oh-l-proline
To be Published
|
|
4JD7
 
 | | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate | | Descriptor: | Proline racemase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate
To be Published
|
|
4JED
 
 | | Crystal structure of glutathione s-transferase mrad2831_1084 (target efi-507060) from methylobacterium radiotolerans jcm 2831, complex with glutathione sulfonate | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of glutathione S-transferase (TARGET EFI-507060) from Methylobacterium radiotolerans
To be Published
|
|
4H19
 
 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502087) from agrobacterium tumefaciens, with bound Mg and d-ribonohydroxamate, ordered loop | | Descriptor: | (2R,3R,4R)-N,2,3,4,5-pentakis(oxidanyl)pentanamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502087) from agrobacterium tumefaciens, with bound Mg and d-ribonohydroxamate, ordered loop
To be Published
|
|
4GP2
 
 | | Crystal structure of ISOPRENOID SYNTHASE A3MSH1 (TARGET EFI-501992) from pyrobaculum calidifontis complexed with DMAPP and Magnesium | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, Polyprenyl synthetase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase from Pyrobaculum Calidifontis
To be Published
|
|
