8CLZ
 
 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (monoclinic form R4mC-2) | | Descriptor: | CHLORIDE ION, Putative L-asparaginase II protein, ZINC ION | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CLY
 
 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (tetragonal form R4tP) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative L-asparaginase II protein, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2BL2
 
 | | The membrane rotor of the V-type ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, SODIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2005-02-25 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Rotor of the Vacuolar-Type Na- ATPase from Enterococcus Hirae
Science, 308, 2005
|
|
2C5F
 
 | | Torpedo californica acetylcholinesterase in complex with a non hydrolysable substrate analogue, 4-oxo-N,N,N-trimethylpentanaminium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Substrate Traffic and Inhibition in Acetylcholinesterase.
Embo J., 25, 2006
|
|
3V47
 
 | | Crystal structure of the N-terminal fragment of zebrafish TLR5 in complex with Salmonella flagellin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Flagellin, ... | | Authors: | Yoon, S.I, Hong, H, Wilson, I.A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis of TLR5-flagellin recognition and signaling.
Science, 335, 2012
|
|
5VYC
 
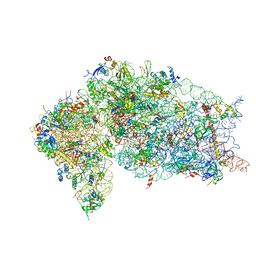 | | Crystal structure of the human 40S ribosomal subunit in complex with DENR-MCT-1. | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Lomakin, I.B, Stolboushkina, E.A, Vaidya, A.T, Garber, M.B, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal Structure of the Human Ribosome in Complex with DENR-MCT-1.
Cell Rep, 20, 2017
|
|
5VJK
 
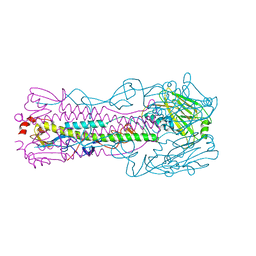 | | Crystal structure of H7 hemagglutinin mutant (V186K, K193T, G228S) from the influenza virus A/Shanghai/2/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Three mutations switch H7N9 influenza to human-type receptor specificity.
PLoS Pathog., 13, 2017
|
|
1J1G
 
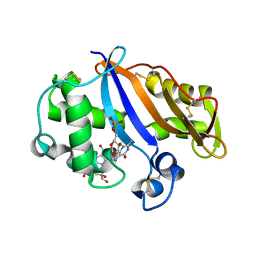 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
2C4H
 
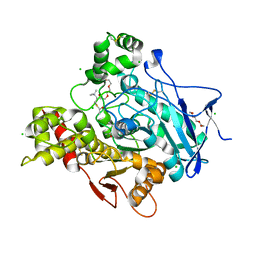 | | Torpedo californica acetylcholinesterase in complex with 500mM acetylthiocholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ACETYL GROUP, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-19 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into substrate traffic and inhibition in acetylcholinesterase.
EMBO J., 25, 2006
|
|
2BN0
 
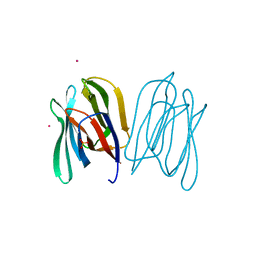 | | Banana Lectin bound to Laminaribiose | | Descriptor: | CADMIUM ION, RIPENING-ASSOCIATED PROTEIN, SULFATE ION, ... | | Authors: | Meagher, J.L, Winter, H.C, Ezell, P, Goldstein, I.J, Stuckey, J.A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Banana Lectin Reveals a Novel Second Sugar Binding Site.
Glycobiology, 15, 2005
|
|
2BTV
 
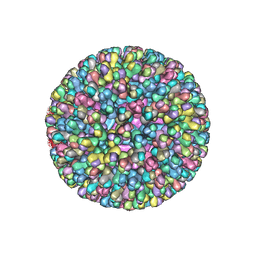 | | ATOMIC MODEL FOR BLUETONGUE VIRUS (BTV) CORE | | Descriptor: | PROTEIN (VP3 CORE PROTEIN), PROTEIN (VP7 CORE PROTEIN) | | Authors: | Grimes, J.M, Burroughs, J.N, Gouet, P, Diprose, J.M, Malby, R, Zientras, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 1998-09-05 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of the bluetongue virus core.
Nature, 395, 1998
|
|
2BTC
 
 | | BOVINE TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2C58
 
 | | Torpedo californica acetylcholinesterase in complex with 20mM acetylthiocholine | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-26 | | Release date: | 2006-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into Substrate Traffic and Inhibition in Acetylcholinesterase.
Embo J., 25, 2006
|
|
4FWG
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with lactacystin | | Descriptor: | Omuralide, open form, PHOSPHATE ION, ... | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8C24
 
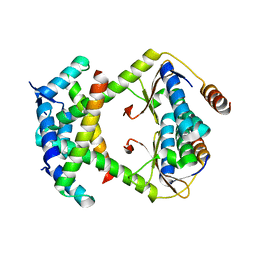 | |
3UN9
 
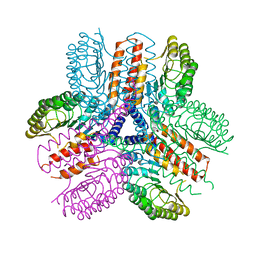 | | Crystal structure of an immune receptor | | Descriptor: | NLR family member X1, PLATINUM (II) ION | | Authors: | Hong, M, Yoon, S.I, Wilson, I.A. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Functional Characterization of the RNA-Binding Element of the NLRX1 Innate Immune Modulator.
Immunity, 36, 2012
|
|
1IS2
 
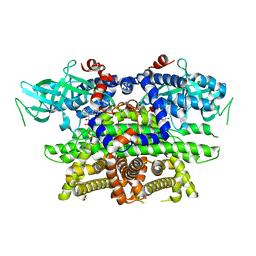 | |
1ITW
 
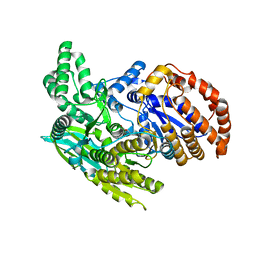 | | Crystal structure of the monomeric isocitrate dehydrogenase in complex with isocitrate and Mn | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase, MANGANESE (II) ION | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-02-12 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Monomeric Isocitrate Dehydrogenase: Evidence of a Protein Monomerization by a Domain Duplication
Structure, 10, 2002
|
|
5EDF
 
 | | Crystal structure of the selenomethionine-substituted iron-regulated protein FrpD from Neisseria meningitidis | | Descriptor: | AZIDE ION, FrpC operon protein, HEXAETHYLENE GLYCOL, ... | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
1J1F
 
 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
2BR7
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, Van Nierop, P, Van Elk, R, Van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha- Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
4O9Y
 
 | | Crystal Structure of TcdA1 | | Descriptor: | TcdA1 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
1IYX
 
 | | Crystal structure of enolase from Enterococcus hirae | | Descriptor: | ENOLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hosaka, T, Meguro, T, Yamato, I, Shirakihara, Y. | | Deposit date: | 2002-09-12 | | Release date: | 2003-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Enterococcus hirae Enolase at 2.8 A Resolution
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0E
 
 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
4FWD
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
