4H4N
 
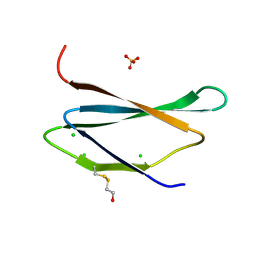 | | 1.1 Angstrom Crystal Structure of Hypothetical Protein BA_2335 from Bacillus anthracis | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-17 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 1.1 Angstrom Crystal Structure of Hypothetical Protein BA_2335 from Bacillus anthracis.
TO BE PUBLISHED
|
|
4IB8
 
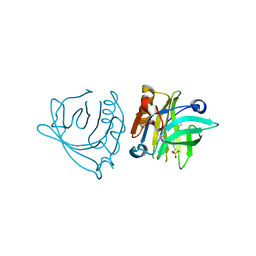 | | Bovine beta-lactoglobulin (isoform A) in complex with dodecyl sulphate (SDS) | | Descriptor: | DODECYL SULFATE, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
4IFG
 
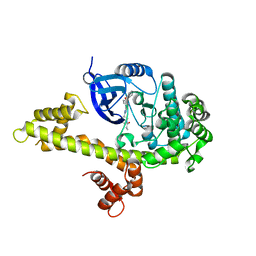 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
4IJ3
 
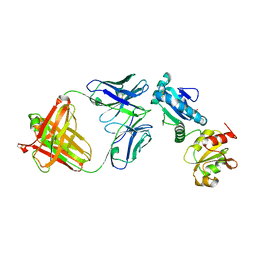 | |
4IB9
 
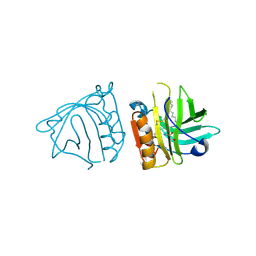 | | Bovine beta-lactoglobulin (isoform B) in complex with dodecyltrimethylammonium (DTAC) | | Descriptor: | DODECANE-TRIMETHYLAMINE, GLYCEROL, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
4IL5
 
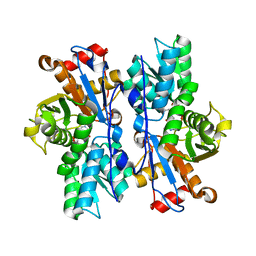 | |
4IH2
 
 | | Crystal structure of kirola (Act d 11) from crystal soaked with 2-aminopurine | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
4IHP
 
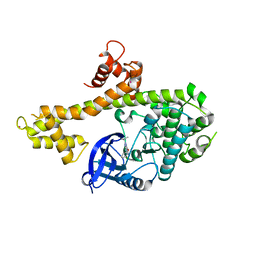 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I.E, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, C.K, Shokat, K.M, Sibley, L.D, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
TO BE PUBLISHED
|
|
4FHP
 
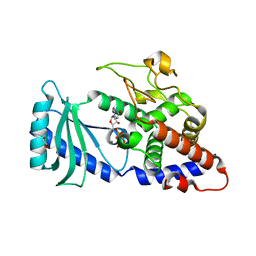 | | Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity - CaUTP bound | | Descriptor: | CALCIUM ION, GLYCEROL, Poly(A) RNA polymerase protein cid1, ... | | Authors: | Lunde, B.M, Magler, I, Meinhart, A. | | Deposit date: | 2012-06-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity.
Nucleic Acids Res., 40, 2012
|
|
4FPQ
 
 | | Structure of a fungal protein | | Descriptor: | Avirulence Effector AvrLm4-7 | | Authors: | Blondeau, K, Blaise, F, Graille, M, Linglin, J, Ollivier, B, Labarde, A, Doizy, A, Daverdin, G, Balesdent, MH, Rouxel, T, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2012-06-22 | | Release date: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the effector AvrLm4-7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins.
Plant J., 83, 2015
|
|
4FWB
 
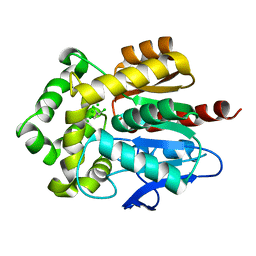 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 in complex with 1, 2, 3 - trichloropropane | | Descriptor: | 1,2,3-trichloropropane, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Kuta Smatanova, I. | | Deposit date: | 2012-06-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HH2
 
 | | Structure of PpsR without the HTH motif from Rb. sphaeroides | | Descriptor: | Transcriptional regulator, PpsR | | Authors: | Winkler, A, Heintz, U, Lindner, R, Reinstein, J, Shoeman, R, Schlichting, I. | | Deposit date: | 2012-10-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GQO
 
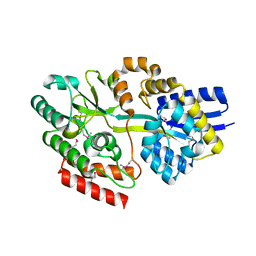 | | 2.1 Angstrom resolution crystal structure of uncharacterized protein lmo0859 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo0859 protein, TRIETHYLENE GLYCOL | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-23 | | Release date: | 2012-09-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of uncharacterized protein lmo0859 from Listeria monocytogenes EGD-e
To be Published
|
|
4HEG
 
 | | Crystal Structure of HIV-1 protease mutants R8Q complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, HIV-1 protease | | Authors: | Zhang, H, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4GYE
 
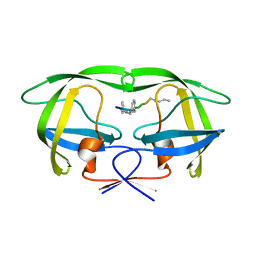 | | MDR 769 HIV-1 Protease in Complex with Reduced P1F | | Descriptor: | P1F peptide, Protease | | Authors: | Dewdney, T.G, Wang, Y, Brunzelle, J, Reiter, S.J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
4H4E
 
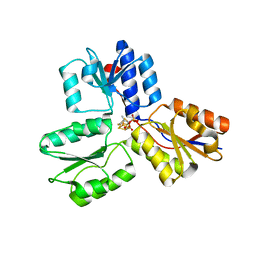 | | IspH in complex with (E)-4-mercapto-3-methylbut-2-enyl diphosphate | | Descriptor: | (2E)-3-methyl-4-sulfanylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Eisenreich, W, Jauch, J, Bacher, A, Groll, M. | | Deposit date: | 2012-09-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Fluoro, Amino, and Thiol Inhibitors Bound to the [Fe(4) S(4) ] Protein IspH.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4HDB
 
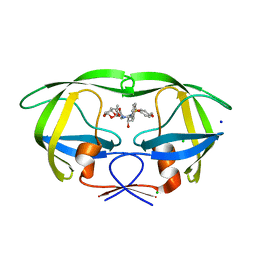 | | Crystal Structure of HIV-1 protease mutants D30N complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Zhang, H, Wang, Y.-F, Shen, C.H, Agniswamy, J, Weber, I.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4HGX
 
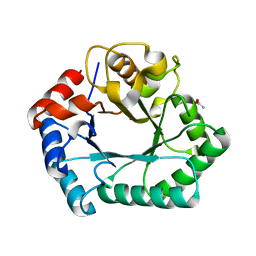 | | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand | | Descriptor: | ACETATE ION, Xylose isomerase domain containing protein, ZINC ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Dorovatovsky, P.V, Rakitina, T.V, Lipkin, A.V, Shumilin, I.A, Minor, W, Popov, V.O. | | Deposit date: | 2012-10-09 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand
To be Published
|
|
4H7Z
 
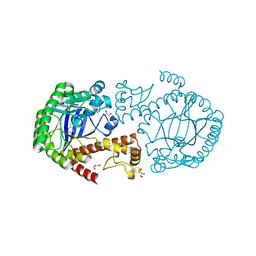 | | tRNA-guanine transglycosylase Y106F, C158V mutant in complex with guanine | | Descriptor: | GLYCEROL, GUANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
4HDF
 
 | | Crystal Structure of HIV-1 protease mutants V82A complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease | | Authors: | Zhang, H, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4IFO
 
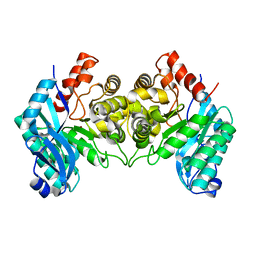 | | 2.50 Angstroms X-ray crystal structure of R51A 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase from Pseudomonas fluorescens | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
4IG2
 
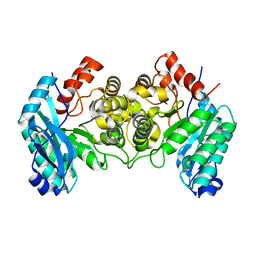 | | 1.80 Angstroms X-ray crystal structure of R51A and R239A heterodimer 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase from Pseudomonas fluorescens | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-12-15 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
6JT4
 
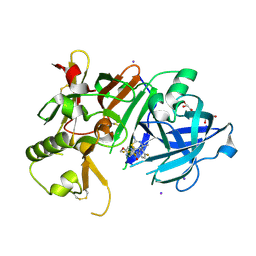 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
4IR4
 
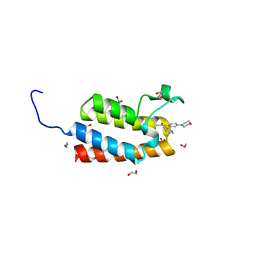 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone (GSK2834113A) | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
4IR5
 
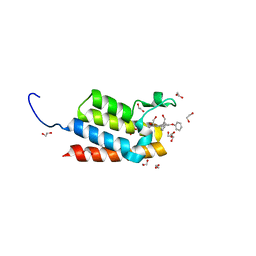 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone (GSK2847449A) | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
