5V8K
 
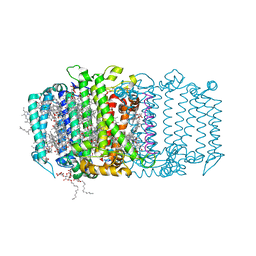 | | Homodimeric reaction center of H. modesticaldum | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, 4,4'-Diaponeurosporene, 8(1)-OH-Chlorophyll aF, ... | | Authors: | Gisriel, C, Sarrou, I, Ferlez, B, Golbeck, J, Redding, K.E, Fromme, R. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a symmetric photosynthetic reaction center-photosystem.
Science, 357, 2017
|
|
1QBA
 
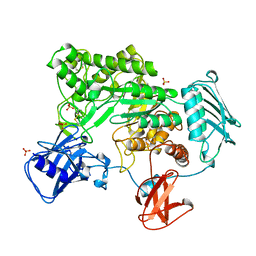 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
8RAS
 
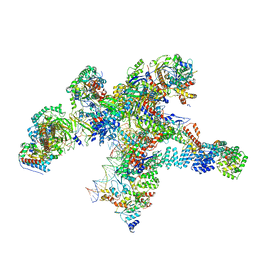 | | Plastid-encoded RNA polymerase transcription elongation complex | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
1QFE
 
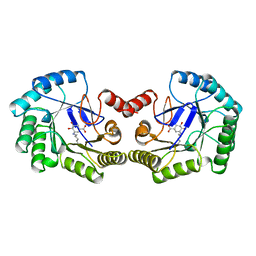 | | THE STRUCTURE OF TYPE I 3-DEHYDROQUINATE DEHYDRATASE FROM SALMONELLA TYPHI | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, PROTEIN (3-DEHYDROQUINATE DEHYDRATASE) | | Authors: | Shrive, A.K, Polikarpov, I, Sawyer, L, Coggins, J.R, Hawkins, A.R. | | Deposit date: | 1999-04-05 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction.
Nat.Struct.Biol., 6, 1999
|
|
6CGI
 
 | | Structure of Salmonella Effector SseK3 | | Descriptor: | Type III secretion system effector protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Chung, I.Y.W, Cygler, M. | | Deposit date: | 2018-02-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Salmonella Effectors SseK1 and SseK3 Target Death Domain Proteins in the TNF and TRAIL Signaling Pathways.
Mol.Cell Proteomics, 18, 2019
|
|
1T9O
 
 | | Crystal Structure of V44G Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
1JDV
 
 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH ADENOSINE AND SULFATE ION | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, ADENOSINE, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
5NO4
 
 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate with uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
7W1C
 
 | | Crystal structure of Klebsiella pneumoniae K1 capsule-specific polysaccharide lyase in a P1 crystal form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, IMIDAZOLE, ... | | Authors: | Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and biological insights into Klebsiella pneumoniae surface polysaccharide degradation by a bacteriophage K1 lyase: implications for clinical use.
J.Biomed.Sci., 29, 2022
|
|
5NFY
 
 | | SARS-CoV nsp10/nsp14 dynamic complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Polyprotein 1ab, ZINC ION | | Authors: | Ferron, F, Gluais, L, Vonrhein, C, Bricogne, G, Canard, B, Imbert, I. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-10 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.382 Å) | | Cite: | Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7W1D
 
 | | Crystal structure of Klebsiella pneumoniae K1 capsule-specific polysaccharide lyase in a C2 crystal form | | Descriptor: | CARBONATE ION, CITRIC ACID, K1 LYASE | | Authors: | Tu, I.F, Ko, T.P, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and biological insights into Klebsiella pneumoniae surface polysaccharide degradation by a bacteriophage K1 lyase: implications for clinical use.
J.Biomed.Sci., 29, 2022
|
|
3H9R
 
 | | Crystal structure of the kinase domain of type I activin receptor (ACVR1) in complex with FKBP12 and dorsomorphin | | Descriptor: | 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, Activin receptor type-1, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Chaikuad, A, Alfano, I, Shrestha, B, Muniz, J.R.C, Petrie, K, Fedorov, O, Phillips, C, Bishop, S, Mahajan, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Bone Morphogenetic Protein Receptor ALK2 and Implications for Fibrodysplasia Ossificans Progressiva.
J.Biol.Chem., 287, 2012
|
|
4F61
 
 | | Tubulin:Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gigant, B, Mignot, I, Knossow, M. | | Deposit date: | 2012-05-14 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Design and characterization of modular scaffolds for tubulin assembly.
J.Biol.Chem., 287, 2012
|
|
5NO2
 
 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5NAV
 
 | |
3LFZ
 
 | | Crystal Structure of Protein MJ1225 from Methanocaldococcus jannaschii, a putative archaeal homolog of g-AMPK. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gomez-Garcia, I, Oyenarte, I, Kortazar, D, Martinez-Cruz, L.A. | | Deposit date: | 2010-01-19 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of protein MJ1225 from Methanocaldococcus jannaschii shows strong conservation of key structural features seen in the eukaryal gamma-AMPK.
J.Mol.Biol., 65, 2010
|
|
6D2V
 
 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
5NA6
 
 | | Structure of Cys-null Se-Met DPP III from Bacteroides thetaiotaomicron | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Sabljic, I, Luic, M. | | Deposit date: | 2017-02-27 | | Release date: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dipeptidyl peptidase III from the human gut symbiont Bacteroides thetaiotaomicron.
PLoS ONE, 12, 2017
|
|
6J83
 
 | | Crystal structure of TleB with NMVT | | Descriptor: | Cytochrome P-450, N-[(2S)-1-hydroxy-3-(1H-indol-3-yl)propan-2-yl]-N~2~-methyl-L-valinamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fei, H, Mori, T, Abe, I. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
1TPG
 
 | | F1-G MODULE PAIR RESIDUES 1-91 (C83S) OF TISSUE-TYPE PLASMINOGEN ACTIVATOR (T-PA) (NMR, 298K, PH2.95, REPRESENTATIVE STRUCTURE) | | Descriptor: | T-PLASMINOGEN ACTIVATOR F1-G | | Authors: | Smith, B.O, Downing, A.K, Driscoll, P.C, Dudgeon, T.J, Campbell, I.D. | | Deposit date: | 1995-06-14 | | Release date: | 1995-09-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure and backbone dynamics of the fibronectin type I and epidermal growth factor-like pair of modules of tissue-type plasminogen activator.
Structure, 3, 1995
|
|
5AQT
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (1S,2R,3R,5R)-3-(hydroxymethyl)-5-(quinazolin-4-ylamino)cyclopentane-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
3MY0
 
 | | Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Serine/threonine-protein kinase receptor R3 | | Authors: | Chaikuad, A, Alfano, I, Cooper, C, Mahajan, P, Daga, N, Sanvitale, C, Fedorov, O, Petrie, K, Savitsky, P, Gileadi, O, Sethi, R, Krojer, T, Muniz, J.R.C, Pike, A.C.W, Vollmar, M, Carpenter, C.P, Ugochukwu, E, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis.
Angiogenesis, 18, 2015
|
|
1FMH
 
 | |
5AQW
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (1S,2R,3R,5R)-3-(hydroxymethyl)-5-(quinazolin-4-ylamino)cyclopentane-1,2-diol, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQM
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, GLYCEROL, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
