1U4Q
 
 | | Crystal Structure of Repeats 15, 16 and 17 of Chicken Brain Alpha Spectrin | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Kusunoki, H, Minasov, G, MacDonald, R.I, Mondragon, A. | | Deposit date: | 2004-07-26 | | Release date: | 2004-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Independent Movement, Dimerization and Stability of Tandem Repeats of Chicken Brain alpha-Spectrin
J.Mol.Biol., 344, 2004
|
|
5FJ6
 
 | | Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
6AW6
 
 | | 1.70A resolution structure of catechol O-methyltransferase (COMT) L136M (rhombohedral form) from Nannospalax galili | | Descriptor: | Catechol O-methyltransferase, SULFATE ION | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
3RXH
 
 | | Crystal structure of Trypsin complexed with 2-(1H-imidazol-4-yl)ethanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
2NDB
 
 | |
1UA5
 
 | |
5FBM
 
 | | Crystal Structure of Histone Like Protein (HLP) from Streptococcus mutans Refined to 1.9 A Resolution | | Descriptor: | DNA-binding protein HU | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, O'Neil, P, Biswas, I. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of histone-like protein from Streptococcus mutans refined to 1.9 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6ESX
 
 | | Caulobacter crescentus Trx1 | | Descriptor: | Thioredoxin 1 | | Authors: | Van Molle, I, Wahni, K, Goemans, C.V, Beaufay, F, Collet, J.F, Messens, J. | | Deposit date: | 2017-10-24 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | An essential thioredoxin is involved in the control of the cell cycle in the bacterium
J. Biol. Chem., 293, 2018
|
|
2N9H
 
 | | Glucose as a nuclease mimic in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2UY9
 
 | | E162A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
3RXF
 
 | | Crystal structure of Trypsin complexed with 4-aminopyridine | | Descriptor: | 4-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXG
 
 | | Crystal structure of Trypsin complexed with 4-aminocyclohexanol | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
1TZV
 
 | | T. maritima NusB, P3121, Form 1 | | Descriptor: | N utilization substance protein B homolog | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
5FRB
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from a pathogenic filamentous fungus Aspergillus fumigatus in complex with a tetrazole-based inhibitor VT-1598 | | Descriptor: | (R)-4-((4-((6-(2-(2,4-difluorophenyl)-1,1-difluoro-2-hydroxy-3-(1H-tetrazol-1-yl)propyl)pyridin-3-yl)ethynyl)phenoxy)methyl)benzonitrile, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2015-12-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of the New Investigational Drug Candidate VT-1598 in Complex with Aspergillus fumigatus Sterol 14 alpha-Demethylase Provides Insights into Its Broad-Spectrum Antifungal Activity.
Antimicrob. Agents Chemother., 61, 2017
|
|
6B30
 
 | | Structure of RORgt in complex with a novel inverse agonist 1 | | Descriptor: | N-[(1R)-1-(4-methoxyphenyl)-2-oxo-2-{[4-(trimethylsilyl)phenyl]amino}ethyl]-N-methyl-3-oxo-2,3-dihydro-1,2-oxazole-5-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of orally efficacious ROR gamma t inverse agonists, part 1: Identification of novel phenylglycinamides as lead scaffolds.
Bioorg. Med. Chem., 26, 2018
|
|
6B1R
 
 | |
3RXC
 
 | | Crystal structure of Trypsin complexed with 2-aminopyridine | | Descriptor: | 2-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
1TQH
 
 | | Covalent Reaction intermediate Revealed in Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est30 | | Descriptor: | Carboxylesterase precursor, PROPYL ACETATE, SULFATE ION | | Authors: | Liu, P, Wang, Y.F, Ewis, H.E, Abdelal, A.T, Lu, C.D, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30.
J.Mol.Biol., 342, 2004
|
|
3RXQ
 
 | | Crystal structure of Trypsin complexed with benzamide (F01 and F05, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
5FP5
 
 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 4- fluorobenzoic acid (AT222) in an alternate binding site. | | Descriptor: | 4-fluorobenzoic acid, ACETYL GROUP, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FP6
 
 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol (AT17833) in an alternate binding site. | | Descriptor: | 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1TYO
 
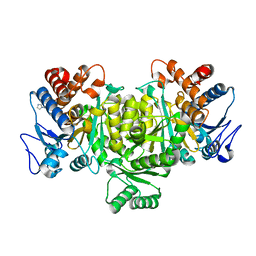 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix in complex with etheno-NADP | | Descriptor: | ETHENO-NADP, isocitrate dehydrogenase | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N, Ladenstein, R. | | Deposit date: | 2004-07-08 | | Release date: | 2005-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
3RXJ
 
 | | Crystal structure of Trypsin complexed with 4-guanidinobenzoic acid | | Descriptor: | 4-carbamimidamidobenzoic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3WBI
 
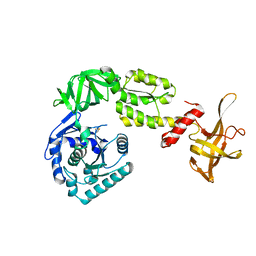 | | Crystal structure analysis of eukaryotic translation initiation factor 5B structure I | | Descriptor: | Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1TZT
 
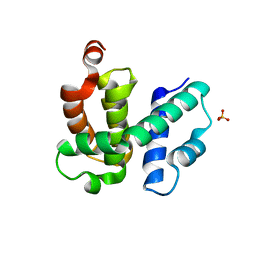 | | T. maritima NusB, P21 | | Descriptor: | N utilization substance protein B homolog, SULFATE ION | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
