6PCR
 
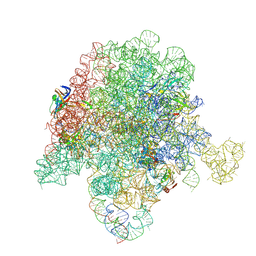 | | E. coli 50S ribosome bound to compound 40o | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl (2-bromopyridin-4-yl)carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
7F1G
 
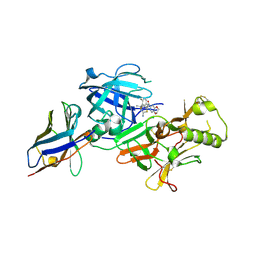 | | BACE2 xaperone complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-2H,3H-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Descriptor: | Beta-secretase 2, N-[3-[(4R,5R,6R)-2-azanyl-5-fluoranyl-4,6-dimethyl-5,6-dihydro-1,3-thiazin-4-yl]-4-fluoranyl-phenyl]-2,3-dihydro-[1,4]dioxino[2,3-c]pyridine-7-carboxamide, XAPERONE | | Authors: | Ueno, T, Matsuoka, E, Asada, N, Yamamoto, S, Kanegawa, N, Ito, M, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2021-06-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Extremely Selective Fused Pyridine-Derived beta-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions.
J.Med.Chem., 64, 2021
|
|
8A2E
 
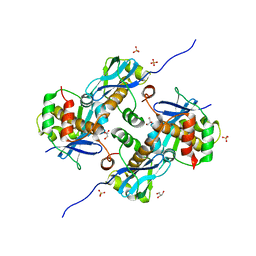 | | Crystal Structure of Human Parechovirus 3 2A protein | | Descriptor: | 2A protein, GLYCEROL, SULFATE ION | | Authors: | von Castelmur, E, Zhu, L, wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family.
To Be Published
|
|
6YHT
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | CITRIC ACID, Conk-C1, SULFATE ION | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
6YHY
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conk-S1 | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
8A5R
 
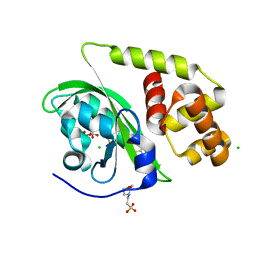 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized and measured in dark. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Light-dependent flavin redox and adduct states control the conformation and DNA-binding activity of the transcription factor EL222.
Nucleic Acids Res., 53, 2025
|
|
8A9F
 
 | |
6PC5
 
 | | E. coli 50S ribosome bound to compounds 46 and VS1 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
7FCW
 
 | | X-ray structure of H2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-07-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7FCU
 
 | | X-ray structure of D2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-07-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7F0Y
 
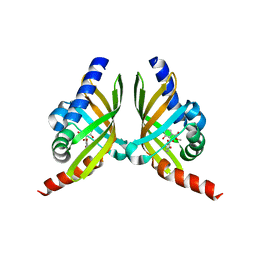 | |
6YFV
 
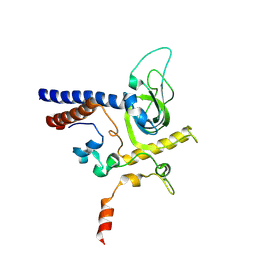 | | Crystal structure of Mtr4-Red1 minimal complex from Chaetomium thermophilum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP dependent RNA helicase (Dob1)-like protein, Red1, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
7F14
 
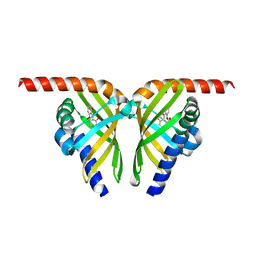 | |
6YRL
 
 | |
7F2G
 
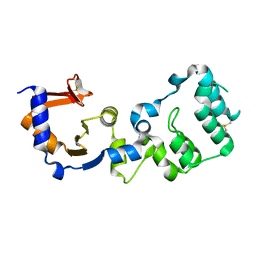 | |
7F13
 
 | | Crystal structure of isomerase Dcr3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dcr3 | | Authors: | Yang, J, Mori, T, Abe, I. | | Deposit date: | 2021-06-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Isomerization Reactions in Fungal Tetrahydroxanthone Biosynthesis and Diversification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7F2H
 
 | |
7F0O
 
 | |
7F10
 
 | |
7EYU
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase SptF-N65T mutant with andiconin D | | Descriptor: | 2-oxoglutarate/Fe(II)-dependent dioxygenase SptF, Andiconin D, FE (II) ION, ... | | Authors: | Tao, H, Mori, T, Abe, I. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular insights into the unusually promiscuous and catalytically versatile Fe(II)/ alpha-ketoglutarate-dependent oxygenase SptF.
Nat Commun, 13, 2022
|
|
7F0Z
 
 | | Crystal structure of NsrQ W31A | | Descriptor: | NsrQ | | Authors: | Yang, J, Mori, T, Abe, I. | | Deposit date: | 2021-06-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Isomerization Reactions in Fungal Tetrahydroxanthone Biosynthesis and Diversification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7F11
 
 | | Crystal structure of NsrQ M128I in complex with substrate analogue 7 | | Descriptor: | NsrQ, methyl 2-[2,6-bis(oxidanyl)phenyl]carbonyl-5-methyl-3,6-bis(oxidanyl)benzoate | | Authors: | Yang, J, Mori, T, Abe, I. | | Deposit date: | 2021-06-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Isomerization Reactions in Fungal Tetrahydroxanthone Biosynthesis and Diversification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7FCB
 
 | | SptF 9 residues truncated mutant | | Descriptor: | 2-oxoglutarate/Fe(II)-dependent dioxygenase SptF, FE (II) ION, N-OXALYLGLYCINE | | Authors: | Hui, T, Mori, T, Abe, I. | | Deposit date: | 2021-07-14 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular insights into the unusually promiscuous and catalytically versatile Fe(II)/ alpha-ketoglutarate-dependent oxygenase SptF.
Nat Commun, 13, 2022
|
|
6PCS
 
 | | E. coli 50S ribosome bound to compound 40e | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl [4-(trifluoromethyl)phenyl]carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
7EQE
 
 | | Crystal Structure of a transcription factor | | Descriptor: | TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
