2MPV
 
 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
8V1I
 
 | | Crystal structure of human mascRNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Skeparnias, I, Zhang, J. | | Deposit date: | 2023-11-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of MALAT1 RNA maturation and mascRNA biogenesis.
Nat.Struct.Mol.Biol., 2024
|
|
1C72
 
 | | TYR115, GLN165 AND TRP209 CONTRIBUTE TO THE 1,2-EPOXY-3-(P-NITROPHENOXY)PROPANE CONJUGATING ACTIVITIES OF GLUTATHIONE S-TRANSFERASE CGSTM1-1 | | Descriptor: | 1-HYDROXY-2-S-GLUTATHIONYL-3-PARA-NITROPHENOXY-PROPANE, PROTEIN (GLUTATHIONE S-TRANSFERASE) | | Authors: | Chern, M.K, Wu, T.C, Hsieh, C.H, Chou, C.C, Liu, L.F, Kuan, I.C, Yeh, Y.H, Hsiao, C.D, Tam, M.F. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tyr115, gln165 and trp209 contribute to the 1, 2-epoxy-3-(p-nitrophenoxy)propane-conjugating activity of glutathione S-transferase cGSTM1-1.
J.Mol.Biol., 300, 2000
|
|
5OBW
 
 | | Mycoplasma genitalium DnaK-NBD in complex with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
1PXK
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)pyrimidin-2-yl]-N'-hydroxyiminoformamide | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-YL]-N'-HYDROXYIMIDOFORMAMIDE | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
5OH8
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Rolipram | | Descriptor: | Cereblon isoform 4, ROLIPRAM, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
2BTC
 
 | | BOVINE TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5NS7
 
 | | Crystal structure of beta-glucosidase BglM-G1 mutant H75R from marine metagenome | | Descriptor: | GLYCEROL, SULFATE ION, beta-glucosidase M - G1 | | Authors: | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
5NZG
 
 | | Crystal structure of UDP-glucose pyrophosphorylase S374W mutant from Leishmania major in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose pyrophosphorylase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
1HTN
 
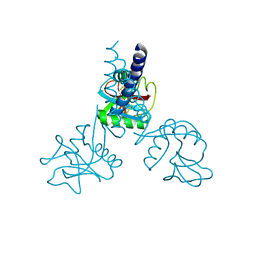 | | HUMAN TETRANECTIN, A TRIMERIC PLASMINOGEN BINDING PROTEIN WITH AN ALPHA-HELICAL COILED COIL | | Descriptor: | CALCIUM ION, TETRANECTIN | | Authors: | Nielsen, B.B, Kastrup, J.S, Rasmussen, H, Holtet, T.L, Graversen, J.H, Etzerodt, M, Thogersen, H.C, Larsen, I.K. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of tetranectin, a trimeric plasminogen-binding protein with an alpha-helical coiled coil.
FEBS Lett., 412, 1997
|
|
2JIM
 
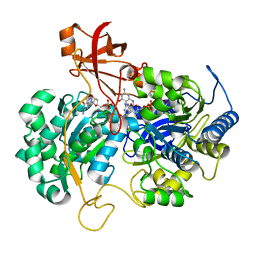 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
1HVW
 
 | |
6C7B
 
 | |
2JM2
 
 | | Structure of the N-terminal subdomain of insulin-like growth factor (IGF) binding protein-6 and its interactions with IGFs | | Descriptor: | Insulin-like growth factor-binding protein 6 | | Authors: | Chandrashekaran, I.R, Yao, S, Wang, C.C, Bansal, P.S, Alewood, P.F, Forbes, B.E, Wallace, J.C, Bach, L.A, Norton, R.S. | | Deposit date: | 2006-09-18 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The N-Terminal Subdomain of Insulin-like Growth Factor (IGF) Binding Protein 6. Structure and Interaction with IGFs
Biochemistry, 46, 2007
|
|
7PZO
 
 | | mite allergen Der p 3 from Dermatophagoides pteronyssinus | | Descriptor: | SULFATE ION, mite allergen Der p 3 | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Mikheeva, O.O, Kostromina, M.A, Lykoshin, D.D, Zayats, E.A, Zavriev, S.K, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural plasticity and thermal stability of the histone-like protein from Spiroplasma melliferum are due to phenylalanine insertions into the conservative scaffold.
J.Biomol.Struct.Dyn., 36, 2018
|
|
6TLE
 
 | | Human Eg5 motor domain mutant E344K | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Garcia-Saez, I, Skoufias, D.A. | | Deposit date: | 2019-12-02 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human Eg5 motor domain mutant E344K
To Be Published
|
|
6TRL
 
 | | Human Eg5 motor domain mutant Y82F | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Kinesin-like protein KIF11, ... | | Authors: | Garcia-Saez, I, Skoufias, D.A. | | Deposit date: | 2019-12-19 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human Eg5 motor domain mutant Y82F
To Be Published
|
|
4YZF
 
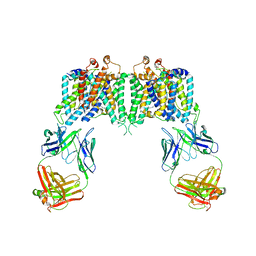 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | Authors: | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-03-25 | | Release date: | 2015-11-04 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
5O60
 
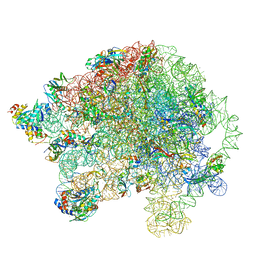 | | Structure of the 50S large ribosomal subunit from Mycobacterium smegmatis | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
1PNJ
 
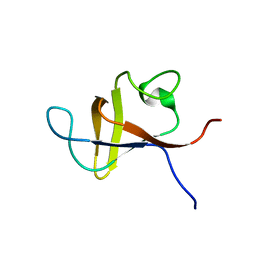 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
7Q3Q
 
 | |
5OAT
 
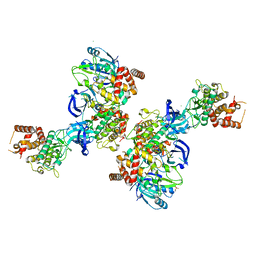 | | PINK1 structure | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein kinase PINK1, mitochondrial-like Protein | | Authors: | Kumar, A, Tamjar, J, Woodroof, H.I, Raimi, O.G, Waddell, A.Y, Peggie, M, Muqit, M.M.K, van Aalten, D.M.F. | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of PINK1 and mechanisms of Parkinson's disease associated mutations.
Elife, 6, 2017
|
|
7Q3R
 
 | |
1YXQ
 
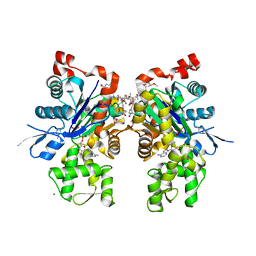 | | Crystal structure of actin in complex with swinholide A | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klenchin, V.A, King, R, Tanaka, J, Marriott, G, Rayment, I. | | Deposit date: | 2005-02-22 | | Release date: | 2005-05-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of swinholide a binding to actin
Chem.Biol., 12, 2005
|
|
5OBY
 
 | | Mycoplasma genitalium DnaK-NBD in complex with AMP-PNP | | Descriptor: | Chaperone protein DnaK, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
