7X7N
 
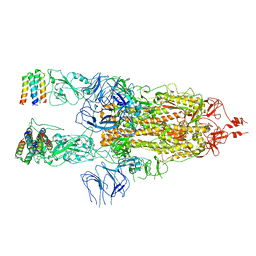 | | 3D model of the 3-RBD up single trimeric spike protein of SARS-CoV2 in the presence of synthetic peptide SIH-5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, Synthetic peptide SIH-5 | | Authors: | Khatri, B, Pramanick, I, Malladi, S.K, Rajmani, R.S, Kumar, S, Ghosh, P, Sengupta, N, Rahisuddin, R, Kumaran, S, Ringe, R.P, Varadarajan, R, Dutta, S, Chatterjee, J. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | A dimeric proteomimetic prevents SARS-CoV-2 infection by dimerizing the spike protein.
Nat.Chem.Biol., 18, 2022
|
|
1JXD
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1T5W
 
 | | HLA-DR1 in complex with a synthetic peptide (AAYSDQATPLLLSPR) | | Descriptor: | 15-mer peptide fragment of Regulatory protein MIG1, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Zavala-Ruiz, Z, Strug, I, Anderson, M.W, Gorski, J, Stern, L.J. | | Deposit date: | 2004-05-05 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Polymorphic Pocket at the P10 Position Contributes to Peptide Binding Specificity in Class II MHC Proteins
Chem.Biol., 11, 2004
|
|
5NS8
 
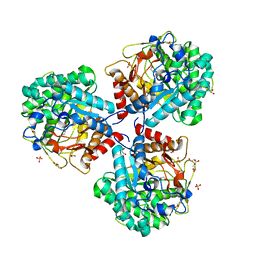 | | Crystal structure of beta-glucosidase BglM-G1 mutant H75R from marine metagenome in complex with inhibitor 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, GLYCEROL, SULFATE ION, ... | | Authors: | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
5O5J
 
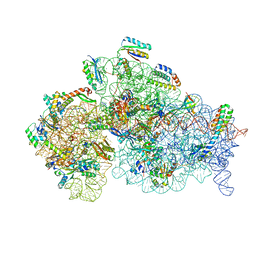 | | Structure of the 30S small ribosomal subunit from Mycobacterium smegmatis | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.451 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
4ADS
 
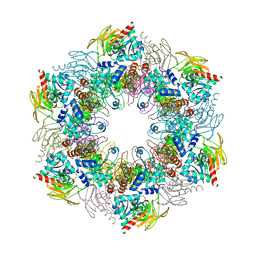 | | Crystal structure of plasmodial PLP synthase complex | | Descriptor: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
2BMZ
 
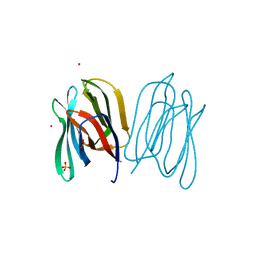 | | Banana Lectin bound to Xyl-b1,3 Man-a-O-Methyl (XM) | | Descriptor: | CADMIUM ION, RIPENING-ASSOCIATED PROTEIN, SULFATE ION, ... | | Authors: | Meagher, J.L, Winter, H.C, Ezell, P, Goldstein, I.J, Stuckey, J.A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Banana Lectin Reveals a Novel Second Sugar Binding Site.
Glycobiology, 15, 2005
|
|
2BMY
 
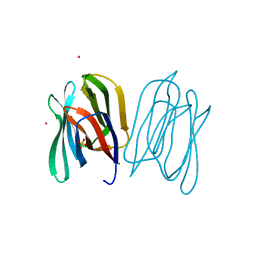 | | Banana Lectin | | Descriptor: | CADMIUM ION, RIPENING-ASSOCIATED PROTEIN, SULFATE ION | | Authors: | Meagher, J.L, Winter, H.C, Ezell, P, Goldstein, I.J, Stuckey, J.A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Banana Lectin Reveals a Novel Second Sugar Binding Site.
Glycobiology, 15, 2005
|
|
1LMS
 
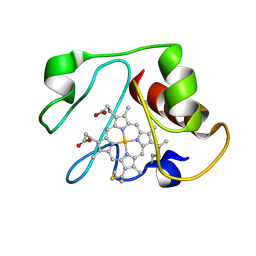 | | Structural model for an alkaline form of ferricytochrome c | | Descriptor: | Cytochrome c, iso-1, HEME C | | Authors: | Assfalg, M, Bertini, I, Dolfi, A, Turano, P, Mauk, A.G, Rosell, F.I, Gray, H.B. | | Deposit date: | 2002-05-02 | | Release date: | 2003-03-18 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural model for an alkaline form of ferricytochrome c
J.Am.Chem.Soc., 125, 2003
|
|
2C5G
 
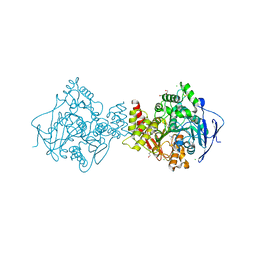 | | Torpedo californica acetylcholinesterase in complex with 20mM thiocholine | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into Substrate Traffic and Inhibition in Acetylcholinesterase.
Embo J., 25, 2006
|
|
1TJL
 
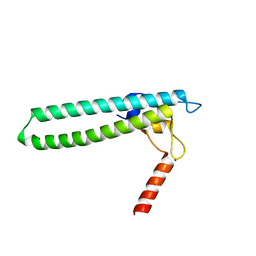 | | Crystal structure of transcription factor DksA from E. coli | | Descriptor: | DnaK suppressor protein, ZINC ION | | Authors: | Perederina, A, Svetlov, V, Vassylyeva, M.N, Artsimovitch, I, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription
Cell(Cambridge,Mass.), 118, 2004
|
|
1H5F
 
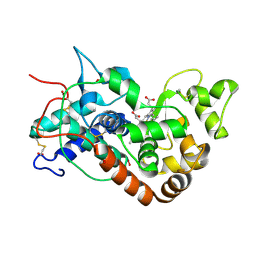 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (22-33% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
1H57
 
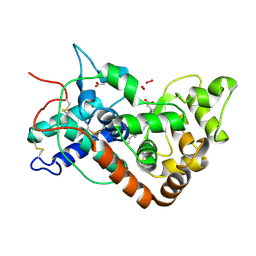 | | Structure of horseradish peroxidase C1A compound III | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J. | | Deposit date: | 2001-05-20 | | Release date: | 2002-06-17 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
5NBC
 
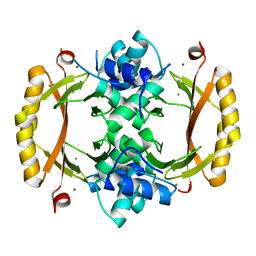 | | Structure of Prokaryotic Transcription Factors | | Descriptor: | Ferric uptake regulation protein, MANGANESE (II) ION, ZINC ION | | Authors: | Perard, J, Carpentier, P, Michaud-Soret, I, Cavazza, C. | | Deposit date: | 2017-03-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role inFrancisella tularensisvirulence.
Commun Biol, 1, 2018
|
|
5NBK
 
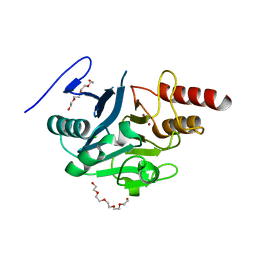 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
1HCH
 
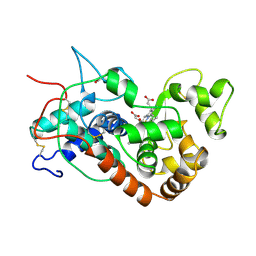 | | Structure of horseradish peroxidase C1A compound I | | Descriptor: | ACETATE ION, CALCIUM ION, OXYGEN ATOM, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-04 | | Release date: | 2002-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1GTZ
 
 | | Structure of STREPTOMYCES COELICOLOR TYPE II DEHYDROQUINASE R23A MUTANT IN COMPLEX WITH DEHYDROSHIKIMATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, 3-DEHYDROSHIKIMATE | | Authors: | Roszak, A.W, Krell, T, Robinson, D.A, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
7Q9W
 
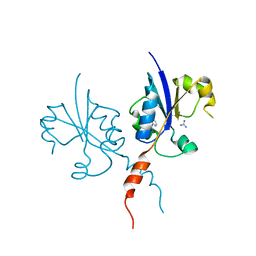 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 4-(aminomethyl)pyridin-2-amine | | Descriptor: | 4-(aminomethyl)pyridin-2-amine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7QA7
 
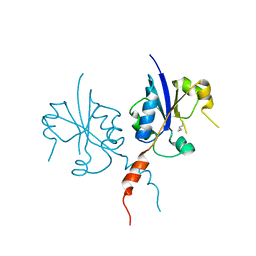 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Cyclopropanemethylamine | | Descriptor: | Cholinephosphate cytidylyltransferase, cyclopropylmethanamine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q9V
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1-(1,3-oxazol-4-yl)methanamine | | Descriptor: | 1,3-oxazol-4-ylmethanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
1SWN
 
 | | CORE-STREPTAVIDIN MUTANT W108F IN COMPLEX WITH BIOTIN AT PH 7.0 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
5N0I
 
 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
4GFA
 
 | | N-terminal coiled-coil dimer of C.elegans SAS-6, crystal form A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Spindle assembly abnormal protein 6 | | Authors: | Erat, M.C, Vakonakis, I. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SWK
 
 | | CORE-STREPTAVIDIN MUTANT W79F IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN, EPI-BIOTIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
