1ECQ
 
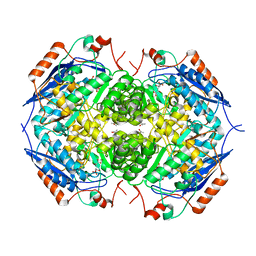 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO 4-DEOXYGLUCARATE | | Descriptor: | 4-DEOXYGLUCARATE, GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1ZDS
 
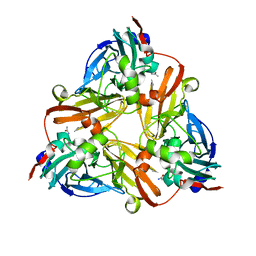 | | Crystal Structure of Met150Gly AfNiR with Acetamide Bound | | Descriptor: | ACETAMIDE, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, MacPherson, I.S, Alexandre, M, Diederix, R.E.M, Canters, G.W, Murphy, M.E.P, Verbeet, M.P. | | Deposit date: | 2005-04-14 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A rearranging ligand enables allosteric control of catalytic activity in copper-containing nitrite reductase.
J.Mol.Biol., 358, 2006
|
|
1ODF
 
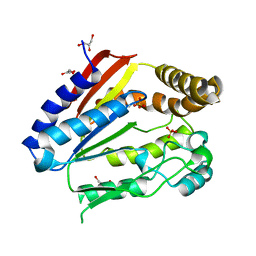 | | Structure of YGR205w protein. | | Descriptor: | GLYCEROL, HYPOTHETICAL 33.3 KDA PROTEIN IN ADE3-SER2 INTERGENIC REGION, SULFATE ION | | Authors: | Li De La Sierra-Gallay, I, Van Tilbeurgh, H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ygr205W Protein from Saccharomyces Cerevisiae: Close Structural Resemblance to E.Coli Pantothenate Kinase
Proteins: Struct.,Funct., Genet., 54, 2004
|
|
4BKX
 
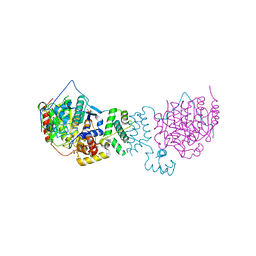 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
1ZML
 
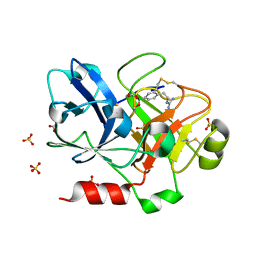 | | Crystal Structure of the Catalytic Domain of Factor XI in complex with (R)-1-(4-(4-(hydroxymethyl)-1,3,2-dioxaborolan-2-yl)phenethyl)guanidine | | Descriptor: | (R)-1-(4-(4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL)PHENETHYL)GUANIDINE, BICARBONATE ION, Coagulation factor XI, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M.J, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R.E, Strickler, J.E. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1Z8L
 
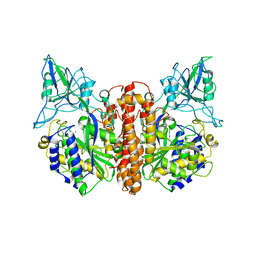 | | Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davis, M.I, Bennett, M.J, Thomas, L.M, Bjorkman, P.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ZDL
 
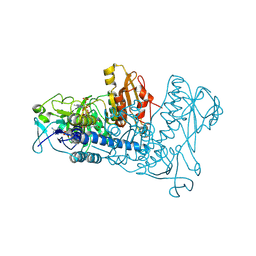 | | Crystal Structure of Mouse Thioredoxin Reductase Type 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 2, ... | | Authors: | Biterova, E.I, Turanov, A.A, Gladyshev, V.N, Barycki, J.J. | | Deposit date: | 2005-04-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1RUI
 
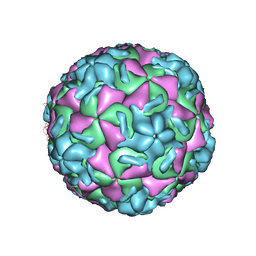 | | RHINOVIRUS 14 MUTANT S1223G COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52084 | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
2YGY
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form II | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL | | Authors: | Campeotto, I, Nelson, A, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2011-04-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathological macromolecular crystallographic data affected by twinning, partial-disorder and exhibiting multiple lattices for testing of data processing and refinement tools.
Sci Rep, 8, 2018
|
|
2YKS
 
 | |
1B2K
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | IODIDE ION, PROTEIN (LYSOZYME) | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-26 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2K6Y
 
 | | Solution structures of apo form PCuA (cis conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2KLK
 
 | | Solution structure of GB1 A34F mutant with RDC and SAXS | | Descriptor: | IMMUNOGLOBULIN G-BINDING PROTEIN G | | Authors: | Wang, J, Zuo, X, Yu, P, Byeon, I.L, Jung, J, Schwieters, C.D, Gronenborn, A.M, Wang, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
2G9O
 
 | | Solution structure of the apo form of the third metal-binding domain of ATP7A protein (Menkes Disease protein) | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
1OJT
 
 | |
4NPO
 
 | | Crystal structure of protein with unknown function from Deinococcus radiodurans at P61 spacegroup | | Descriptor: | ACETATE ION, ACETYL GROUP, GLYCEROL, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Shabalin, I.G, Shumilin, I.A, Dorovatovsky, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of protein with unknown function at P61 spacegroup
To be Published
|
|
2KDI
 
 | | Solution structure of a Ubiquitin/UIM fusion protein | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein 27 fusion protein | | Authors: | Sgourakis, N.G, Patel, M.M, Garcia, A.E, Makhatadze, G.I, McCallum, S.A. | | Deposit date: | 2009-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Conformational Dynamics and Structural Plasticity Play Critical Roles in the Ubiquitin Recognition of a UIM Domain.
J.Mol.Biol., 396, 2010
|
|
1ZGB
 
 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (R)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5R)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
2KFB
 
 | | The structure of the cataract causing P23T mutant of human gamma-D crystallin | | Descriptor: | Gamma-crystallin D | | Authors: | Jung, J, Byeon, I.L, Wang, Y, King, J, Gronenborn, A.M. | | Deposit date: | 2009-02-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the cataract-causing P23T mutant of human gammaD-crystallin exhibits distinctive local conformational and dynamic changes.
Biochemistry, 48, 2009
|
|
1OS2
 
 | | Ternary enzyme-product-inhibitor complexes of human MMP12 | | Descriptor: | ACETATE ION, ACETOHYDROXAMIC ACID, AZIDE ION, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Structures of Binary and Ternary Enzyme-Product-Inhibitor Complexes of Matrix Metalloproteinases
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
1JCT
 
 | | Glucarate Dehydratase, N341L mutant Orthorhombic Form | | Descriptor: | D-GLUCARATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-11 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
3E7C
 
 | | Glucocorticoid Receptor LBD bound to GSK866 | | Descriptor: | 5-amino-N-[(2S)-2-({[(2,6-dichlorophenyl)carbonyl](ethyl)amino}methyl)-3,3,3-trifluoro-2-hydroxypropyl]-1-(4-fluorophenyl)-1H-pyrazole-4-carboxamide, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Madauss, K.P, Williams, S.P, Mclay, I, Stewart, E.L, Bledsoe, R.K. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The first X-ray crystal structure of the glucocorticoid receptor bound to a non-steroidal agonist.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2KL2
 
 | | NMR solution structure of A2LD1 (gi:13879369) | | Descriptor: | AIG2-like domain-containing protein 1 | | Authors: | Pedrini, B, Serrano, P, Mohanty, B, Geralt, M, Herrmann, T, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1Z0G
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
3AFF
 
 | | Crystal structure of the HsaA monooxygenase from M. tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Strynadka, N, Eltis, L.D. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
