1E28
 
 | | Nonstandard peptide binding of HLA-B*5101 complexed with HIV immunodominant epitope KM2(TAFTIPSI) | | 分子名称: | BETA-2 MICROGLOBULIN LIGHT CHAIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HEAVY CHAIN, PEPTIDE | | 著者 | Maenaka, K, Maenaka, T, Tomiyama, H, Takiguchi, M, Stuart, D.I, Jones, E.Y. | | 登録日 | 2000-05-18 | | 公開日 | 2000-09-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
J Immunol., 165, 2000
|
|
7Q87
 
 | | Carboxypeptidase T with (S)-3-phenyllactic acid | | 分子名称: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, CALCIUM ION, Carboxypeptidase T, ... | | 著者 | Timofeev, V.I, Akparov, V.K, Shevtsov, M.B, Kuranova, I.P. | | 登録日 | 2021-11-10 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Carboxypeptidase T with (S)-3-phenyllactic acid
To Be Published
|
|
7VUS
 
 | | Crystal structure of AlleyCat9 with 5-nitro-benzotriazole | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1H-benzotriazole, AlleyCat, ... | | 著者 | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | 登録日 | 2021-11-04 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUR
 
 | | Crystal structure of AlleyCat9 with calcium but no inhibitor | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, AlleyCat, CALCIUM ION | | 著者 | Margheritis, E, Takahashi, K, Korendovych, I.V, Tame, J.R.H. | | 登録日 | 2021-11-04 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | NMR-guided directed evolution.
Nature, 610, 2022
|
|
5KUY
 
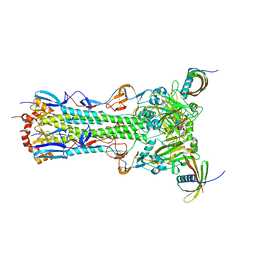 | |
7VUU
 
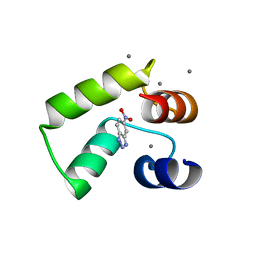 | | Crystal structure of AlleyCat10 with inhibitor | | 分子名称: | 5-nitro-1H-benzotriazole, AlleyCat, CALCIUM ION | | 著者 | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | 登録日 | 2021-11-04 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | NMR-guided directed evolution.
Nature, 610, 2022
|
|
8T1C
 
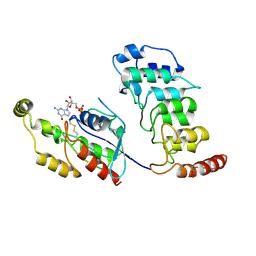 | | Cryo-EM structure of human TRPV4 ankyrin repeat domain in complex with GTPase RhoA | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4-Enhanced green fluorescent protein chimera | | 著者 | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | 登録日 | 2023-06-02 | | 公開日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
6GZH
 
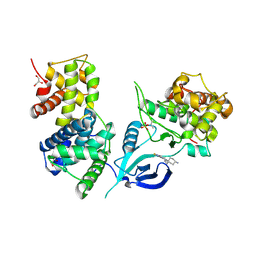 | | Crystal Structure of Human CDK9/cyclinT1 with A86 | | 分子名称: | Cyclin-T1, Cyclin-dependent kinase 9, GLYCEROL, ... | | 著者 | Ben-neriah, Y, Venkatachalam, A, Minzel, W, Fink, A, Snir-Alkalay, I, Vacca, J. | | 登録日 | 2018-07-04 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Small Molecules Co-targeting CKI alpha and the Transcriptional Kinases CDK7/9 Control AML in Preclinical Models.
Cell, 175, 2018
|
|
7VUT
 
 | | Crystal structure of AlleyCat10 | | 分子名称: | AlleyCat10, CALCIUM ION | | 著者 | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | 登録日 | 2021-11-04 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7SH1
 
 | | Class II UvrA protein - Ecm16 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Excinuclease ABC subunit UvrA, ... | | 著者 | Grade, P, Erlandson, A, Ullah, A, Mathews, I.I, Chen, X, Kim, C.-Y, Mera, P.E. | | 登録日 | 2021-10-07 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi.
Sci Rep, 13, 2023
|
|
5OGA
 
 | | Structure of minimal i-motif domain | | 分子名称: | DNA (5'-D(*TP*(DCP)P*GP*TP*TP*CP*(DCP)P*GP*TP*TP*TP*TP*TP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | 著者 | Mir, B, Serrano, I, Buitrago, D, Orozco, M, Escaja, N, Gonzalez, C. | | 登録日 | 2017-07-12 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Prevalent Sequences in the Human Genome Can Form Mini i-Motif Structures at Physiological pH.
J. Am. Chem. Soc., 139, 2017
|
|
7SEH
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403QdLtL) | | 分子名称: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | 登録日 | 2021-09-30 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
7SEI
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403Q) | | 分子名称: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | 登録日 | 2021-09-30 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
6WGI
 
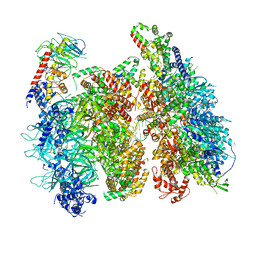 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | 分子名称: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | 著者 | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | 登録日 | 2020-04-05 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1AJF
 
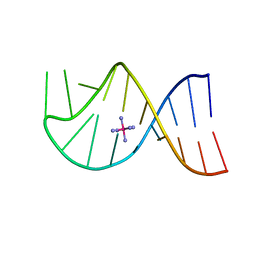 | |
2NNP
 
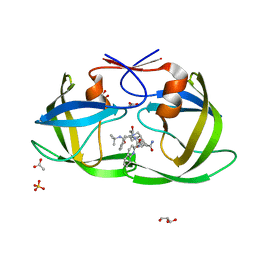 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-24 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2YM8
 
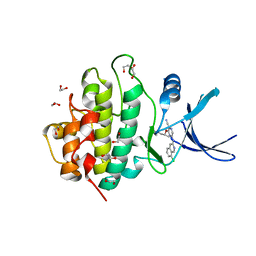 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | 分子名称: | (R)-5-(8-CHLOROISOQUINOLIN-3-YLAMINO)-3-(1-(DIMETHYLAMINO)PROPAN-2-YLOXY)PYRAZINE-2-CARBONITRILE, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | 著者 | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | 登録日 | 2011-06-06 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
1TL5
 
 | | Solution structure of apoHAH1 | | 分子名称: | Copper transport protein ATOX1 | | 著者 | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | 登録日 | 2004-06-09 | | 公開日 | 2004-10-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
6JFD
 
 | |
2NEF
 
 | | HIV-1 NEF (REGULATORY FACTOR), NMR, 40 STRUCTURES | | 分子名称: | NEGATIVE FACTOR (F-PROTEIN) | | 著者 | Grzesiek, S, Bax, A, Clore, G.M, Gronenborn, A.M, Hu, J.S, Kaufman, J, Palmer, I, Stahl, S.J, Tjandra, N, Wingfield, P.T. | | 登録日 | 1997-02-12 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined solution structure and backbone dynamics of HIV-1 Nef.
Protein Sci., 6, 1997
|
|
6JFA
 
 | |
6JFF
 
 | |
6T6M
 
 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 5.5 | | 分子名称: | GLYCEROL, HISTIDINE, Orange carotenoid-binding protein, ... | | 著者 | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | 登録日 | 2019-10-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
1B5P
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE DOUBLE MUTANT 1 | | 分子名称: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | 著者 | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | 登録日 | 1999-01-07 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
6JFE
 
 | |
